A step-by-step tutorial for fine-tuning SAM2 for custom segmentation tasks
SAM2 (Segment Anything 2) is a new model by Meta aiming to segment anything in an image without being limited to specific classes or domains. What makes this model unique is the scale of data on which it was trained: 11 million images, and 11 billion masks. This extensive training makes SAM2 a powerful starting point for training on new image segmentation tasks.
The question you might ask is if SAM can segment anything why do we even need to retrain it? The answer is that SAM is very good at common objects but can perform rather poorly on rare or domain-specific tasks.
However, even in cases where SAM gives insufficient results, it is still possible to significantly improve the model’s ability by fine-tuning it on new data. In many cases, this will take less training data and give better results then training a model from scratch.
This tutorial demonstrates how to fine-tune SAM2 on new data in just 60 lines of code (excluding comments and imports).
The full training script of the can be found in:

How Segment Anything works
The main way SAM works is by taking an image and a point in the image and predicting the mask of the segment that contains the point. This approach enables full image segmentation without human intervention and with no limits on the classes or types of segments (as discussed in a previous post).
The procedure for using SAM for full image segmentation:
- Select a set of points in the image
- Use SAM to predict the segment containing each point
- Combine the resulting segments into a single map
While SAM can also utilize other inputs like masks or bounding boxes, these are mainly relevant for interactive segmentation involving human input. For this tutorial, we’ll focus on fully automatic segmentation and will only consider single points input.
More details on the model are available on the project website.
Downloading SAM2 and setting environment
The SAM2 can be downloaded from:
If you don’t want to copy the training code, you can also download my forked version that already contains the TRAIN.py script.
Follow the installation instructions on the github repository.
In general, you need Python >=3.11 and PyTorch.
In addition, we will use OpenCV this can be installed using:
pip install opencv-python
Downloading pre-trained model
You also need to download the pre-trained model from:
https://github.com/facebookresearch/segment-anything-2?tab=readme-ov-file#download-checkpoints
There are several models you can choose from all compatible with this tutorial. I recommend using the small model which is the fastest to train.
Downloading training data
The next step is to download the dataset that will be used to fine-tune the model. For this tutorial, we will use the LabPics1 dataset for segmenting materials and liquids. You can download the dataset from this URL:
https://zenodo.org/records/3697452/files/LabPicsV1.zip?download=1
Preparing the data reader
The first thing we need to write is the data reader. This will read and prepare the data for the net.
The data reader needs to produce:
- An image
- Masks of all the segments in the image.
- And a random point inside each mask
Lets start by loading dependencies:
import numpy as np
import torch
import cv2
import os
from sam2.build_sam import build_sam2
from sam2.sam2_image_predictor import SAM2ImagePredictor
Next we list all the images in the dataset:
data_dir=r"LabPicsV1//" # Path to LabPics1 dataset folder
data=[] # list of files in dataset
for ff, name in enumerate(os.listdir(data_dir+"Simple/Train/Image/")): # go over all folder annotation
data.append({"image":data_dir+"Simple/Train/Image/"+name,"annotation":data_dir+"Simple/Train/Instance/"+name[:-4]+".png"})
Now for the main function that will load the training batch. The training batch includes: One random image, all the segmentation masks belong to this image, and a random point in each mask:
def read_batch(data): # read random image and its annotaion from the dataset (LabPics)
# select image
ent = data[np.random.randint(len(data))] # choose random entry
Img = cv2.imread(ent["image"])[...,::-1] # read image
ann_map = cv2.imread(ent["annotation"]) # read annotation
# resize image
r = np.min([1024 / Img.shape[1], 1024 / Img.shape[0]]) # scalling factor
Img = cv2.resize(Img, (int(Img.shape[1] * r), int(Img.shape[0] * r)))
ann_map = cv2.resize(ann_map, (int(ann_map.shape[1] * r), int(ann_map.shape[0] * r)),interpolation=cv2.INTER_NEAREST)
# merge vessels and materials annotations
mat_map = ann_map[:,:,0] # material annotation map
ves_map = ann_map[:,:,2] # vessel annotaion map
mat_map[mat_map==0] = ves_map[mat_map==0]*(mat_map.max()+1) # merged map
# Get binary masks and points
inds = np.unique(mat_map)[1:] # load all indices
points= []
masks = []
for ind in inds:
mask=(mat_map == ind).astype(np.uint8) # make binary mask
masks.append(mask)
coords = np.argwhere(mask > 0) # get all coordinates in mask
yx = np.array(coords[np.random.randint(len(coords))]) # choose random point/coordinate
points.append([[yx[1], yx[0]]])
return Img,np.array(masks),np.array(points), np.ones([len(masks),1])
The first part of this function is choosing a random image and loading it:
ent = data[np.random.randint(len(data))] # choose random entry
Img = cv2.imread(ent["image"])[...,::-1] # read image
ann_map = cv2.imread(ent["annotation"]) # read annotation
Note that OpenCV reads images as BGR while SAM expects images as RGB, using […,::-1] to change the image from BGR to RGB.
Note that OpenCV reads images as BGR while SAM expects RGB images. By using […,::-1] we change the image from BGR to RGB.
SAM expects the image size to not exceed 1024, so we are going to resize the image and the annotation map to this size.
r = np.min([1024 / Img.shape[1], 1024 / Img.shape[0]]) # scalling factor
Img = cv2.resize(Img, (int(Img.shape[1] * r), int(Img.shape[0] * r)))
ann_map = cv2.resize(ann_map, (int(ann_map.shape[1] * r), int(ann_map.shape[0] * r)),interpolation=cv2.INTER_NEAREST)
An important point here is that when resizing the annotation map (ann_map) we use INTER_NEAREST mode (nearest neighbors). In the annotation map, each pixel value is the index of the segment it belongs to. As a result, it’s important to use resizing methods that do not introduce new values to the map.
The next block is specific to the format of the LabPics1 dataset. The annotation map (ann_map) contains a segmentation map for the vessels in the image in one channel, and another map for the materials annotation in a different channel. We going to merge them into a single map.
mat_map = ann_map[:,:,0] # material annotation map
ves_map = ann_map[:,:,2] # vessel annotaion map
mat_map[mat_map==0] = ves_map[mat_map==0]*(mat_map.max()+1) # merged map
What this gives us is a a map (mat_map) in which the value of each pixel is the index of the segment to which it belongs (for example: all cells with value 3 belong to segment 3). We want to transform this into a set of binary masks (0/1) where each mask corresponds to a different segment. In addition, from each mask, we want to extract a single point.
inds = np.unique(mat_map)[1:] # list of all indices in map
points= [] # list of all points (one for each mask)
masks = [] # list of all masks
for ind in inds:
mask = (mat_map == ind).astype(np.uint8) # make binary mask for index ind
masks.append(mask)
coords = np.argwhere(mask > 0) # get all coordinates in mask
yx = np.array(coords[np.random.randint(len(coords))]) # choose random point/coordinate
points.append([[yx[1], yx[0]]])
return Img,np.array(masks),np.array(points), np.ones([len(masks),1])
This is it! We got the image (Img), a list of binary masks corresponding to segments in the image (masks), and for each mask the coordinate of a single point inside the mask (points).

Loading the SAM model
Now lets load the net:
sam2_checkpoint = "sam2_hiera_small.pt" # path to model weight
model_cfg = "sam2_hiera_s.yaml" # model config
sam2_model = build_sam2(model_cfg, sam2_checkpoint, device="cuda") # load model
predictor = SAM2ImagePredictor(sam2_model) # load net
First, we set the path to the model weights in: sam2_checkpoint parameter. We downloaded the weights earlier from here. “sam2_hiera_small.pt” refer to the small model but the code will work for any model you choose. Whichever model you choose you need to set the corresponding config file in the model_cfg parameter. The config files are already located in the sub folder “sam2_configs/” of the main repository.
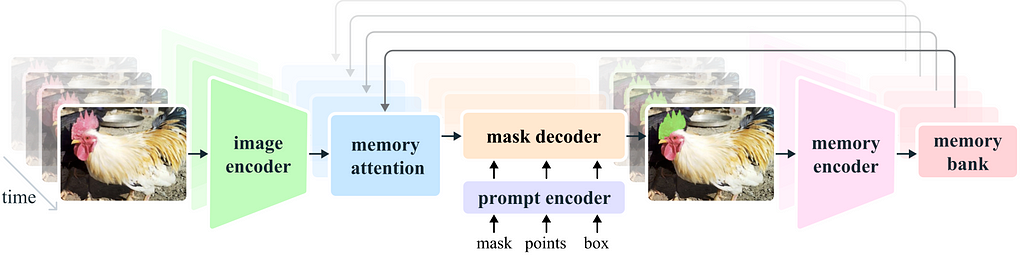
Segment Anything General structure
Before setting training parameters we need to understand the basic structure of the SAM model.
SAM is composed of three parts:
1) Image encoder, 2) Prompt encoder, 3) Mask decoder.
The image encoder is responsible for processing the image and creating the embedding that represents the image. This part consists of a VIT transformer and is the largest component of the net. We usually don’t want to train it, as it already gives good representation and training will demand lots of resources.
The prompt encoder processes the additional input to the net, in our case the input point.
The mask decoder takes the output of the image encoder and prompt encoder and produces the final segmentation masks. In general, we want to train only the mask decoder and maybe the prompt encoder. These parts are lightweight and can be fine-tuned fast with a modest GPU.
Setting training parameters:
We can enable the training of the mask decoder and prompt encoder by setting:
predictor.model.sam_mask_decoder.train(True) # enable training of mask decoder
predictor.model.sam_prompt_encoder.train(True) # enable training of prompt encoder
Next, we define the standard adamW optimizer:
optimizer=torch.optim.AdamW(params=predictor.model.parameters(),lr=1e-5,weight_decay=4e-5)
We also going to use mixed precision training which is just a more memory-efficient training strategy:
scaler = torch.cuda.amp.GradScaler() # set mixed precision
Main training loop
Now lets build the main training loop. The first part is reading and preparing the data:
for itr in range(100000):
with torch.cuda.amp.autocast(): # cast to mix precision
image,mask,input_point, input_label = read_batch(data) # load data batch
if mask.shape[0]==0: continue # ignore empty batches
predictor.set_image(image) # apply SAM image encoder to the image
First we cast the data to mix precision for efficient training:
with torch.cuda.amp.autocast():
Next, we use the reader function we created earlier to read training data:
image,mask,input_point, input_label = read_batch(data)
We take the image we loaded and pass it through the image encoder (the first part of the net):
predictor.set_image(image)
Next, we process the input points using the net prompt encoder:
mask_input, unnorm_coords, labels, unnorm_box = predictor._prep_prompts(input_point, input_label, box=None, mask_logits=None, normalize_coords=True)
sparse_embeddings, dense_embeddings = predictor.model.sam_prompt_encoder(points=(unnorm_coords, labels),boxes=None,masks=None,)
Note that in this part we can also input boxes or masks but we are not going to use these options.
Now that we encoded both the prompt (points) and the image we can finally predict the segmentation masks:
batched_mode = unnorm_coords.shape[0] > 1 # multi mask prediction
high_res_features = [feat_level[-1].unsqueeze(0) for feat_level in predictor._features["high_res_feats"]]
low_res_masks, prd_scores, _, _ = predictor.model.sam_mask_decoder(image_embeddings=predictor._features["image_embed"][-1].unsqueeze(0),image_pe=predictor.model.sam_prompt_encoder.get_dense_pe(),sparse_prompt_embeddings=sparse_embeddings,dense_prompt_embeddings=dense_embeddings,multimask_output=True,repeat_image=batched_mode,high_res_features=high_res_features,)
prd_masks = predictor._transforms.postprocess_masks(low_res_masks, predictor._orig_hw[-1])# Upscale the masks to the original image resolution
The main part in this code is the model.sam_mask_decoder which runs the mask_decoder part of the net and generates the segmentation masks (low_res_masks) and their scores (prd_scores).
These masks are in lower resolution than the original input image and are resized to the original input size in the postprocess_masks function.
This gives us the final prediction of the net: 3 segmentation masks (prd_masks) for each input point we used and the masks scores (prd_scores). prd_masks contains 3 predicted masks for each input point but we only going to use the first mask for each point. prd_scores contains a score of how good the net thinks each mask is (or how sure it is in the prediction).
Loss functions
Segmentation loss
Now we have the net predictions we can calculate the loss. First, we calculate the segmentation loss, which means how good the predicted mask is compared to the ground true mask. For this, we use the standard cross entropy loss.
First we need to convert prediction masks (prd_mask) from logits into probabilities using the sigmoid function:
prd_mask = torch.sigmoid(prd_masks[:, 0])# Turn logit map to probability map
Next we convert the ground truth mask into a torch tensor:
prd_mask = torch.sigmoid(prd_masks[:, 0])# Turn logit map to probability map
Finally, we calculate the cross entropy loss (seg_loss) manually using the ground truth (gt_mask) and predicted probability maps (prd_mask):
seg_loss = (-gt_mask * torch.log(prd_mask + 0.00001) - (1 - gt_mask) * torch.log((1 - prd_mask) + 0.00001)).mean() # cross entropy loss
(we add 0.0001 to prevent the log function from exploding for zero values).
Score loss (optional)
In addition to the masks, the net also predicts the score for how good each predicted mask is. Training this part is less important but can be useful . To train this part we need to first know what is the true score of each predicted mask. Meaning, how good the predicted mask actually is. We are going to do it by comparing the GT mask and the corresponding predicted mask using intersection over union (IOU) metrics. IOU is simply the overlap between the two masks, divided by the combined area of the two masks. First, we calculate the intersection between the predicted and GT mask (the area in which they overlap):
inter = (gt_mask * (prd_mask > 0.5)).sum(1).sum(1)
We use threshold (prd_mask > 0.5) to turn the prediction mask from probability to binary mask.
Next, we get the IOU by dividing the intersection by the combined area (union) of the predicted and gt masks:
iou = inter / (gt_mask.sum(1).sum(1) + (prd_mask > 0.5).sum(1).sum(1) - inter)
We going to use the IOU as the true score for each mask, and get the score loss as the absolute difference between the predicted scores and the IOU we just calculated.
score_loss = torch.abs(prd_scores[:, 0] - iou).mean()
Finally, we merge the segmentation loss and score loss (giving much higher weight to the first):
loss = seg_loss+score_loss*0.05 # mix losses
Final step: Backpropogation and saving model
Once we get the loss everything is completely standard. We calculate backpropogation and update weights using the optimizer we made earlier:
predictor.model.zero_grad() # empty gradient
scaler.scale(loss).backward() # Backpropogate
scaler.step(optimizer)
scaler.update() # Mix precision
We also want to save the trained model once every 1000 steps:
if itr%1000==0: torch.save(predictor.model.state_dict(), "model.torch") # save model
Since we already calculated the IOU we can display it as a moving average to see how well the model prediction are improving over time:
if itr==0: mean_iou=0
mean_iou = mean_iou * 0.99 + 0.01 * np.mean(iou.cpu().detach().numpy())
print("step)",itr, "Accuracy(IOU)=",mean_iou)
And that it, we have trained/ fine-tuned the Segment-Anything 2 in less than 60 lines of code (not including comments and imports). After about 25,000 steps you should see major improvement .
The model will be saved to “model.torch”.
You can find the full training code at:
To see how to load and use the model we just trained check the next section.
Inference: Loading and using the trained model:
Now that the model as been fine-tuned, let’s use it to segment an image.
We going to do this using the following steps:
- Load the model we just trained.
- Give the model an image and a bunch of random points. For each point the net will predict the segment mask that contain this point and a score.
- Take these masks and stitch them together into one segmentation map.
The full code for doing that is available at:
First, we load the dependencies and cast the weights to float16 this makes the model much faster to run (only possible for inference).
import numpy as np
import torch
import cv2
from sam2.build_sam import build_sam2
from sam2.sam2_image_predictor import SAM2ImagePredictor
# use bfloat16 for the entire script (memory efficient)
torch.autocast(device_type="cuda", dtype=torch.bfloat16).__enter__()
Next, we load a sample image and a mask of the image region we want to segment (download image/mask):
image_path = r"sample_image.jpg" # path to image
mask_path = r"sample_mask.png" # path to mask, the mask will define the image region to segment
def read_image(image_path, mask_path): # read and resize image and mask
img = cv2.imread(image_path)[...,::-1] # read image as rgb
mask = cv2.imread(mask_path,0) # mask of the region we want to segment
# Resize image to maximum size of 1024
r = np.min([1024 / img.shape[1], 1024 / img.shape[0]])
img = cv2.resize(img, (int(img.shape[1] * r), int(img.shape[0] * r)))
mask = cv2.resize(mask, (int(mask.shape[1] * r), int(mask.shape[0] * r)),interpolation=cv2.INTER_NEAREST)
return img, mask
image,mask = read_image(image_path, mask_path)
Sample 30 random points inside the region we want to segment:
num_samples = 30 # number of points/segment to sample
def get_points(mask,num_points): # Sample points inside the input mask
points=[]
for i in range(num_points):
coords = np.argwhere(mask > 0)
yx = np.array(coords[np.random.randint(len(coords))])
points.append([[yx[1], yx[0]]])
return np.array(points)
input_points = get_points(mask,num_samples)
Load the standard SAM model (same as in training)
# Load model you need to have pretrained model already made
sam2_checkpoint = "sam2_hiera_small.pt"
model_cfg = "sam2_hiera_s.yaml"
sam2_model = build_sam2(model_cfg, sam2_checkpoint, device="cuda")
predictor = SAM2ImagePredictor(sam2_model)
Next, Load the weights of the model we just trained (model.torch):
predictor.model.load_state_dict(torch.load("model.torch"))
Run the fine-tuned model to predict a segmentation mask for every point we selected earlier:
with torch.no_grad(): # prevent the net from caclulate gradient (more efficient inference)
predictor.set_image(image) # image encoder
masks, scores, logits = predictor.predict( # prompt encoder + mask decoder
point_coords=input_points,
point_labels=np.ones([input_points.shape[0],1])
)
Now we have a list of predicted masks and their scores. We want to somehow stitch them into a single consistent segmentation map. However, many of the masks overlap and might be inconsistent with each other.
The approach to stitching is simple:
First we will sort the predicted masks according to their predicted scores:
masks=masks[:,0].astype(bool)
shorted_masks = masks[np.argsort(scores[:,0])][::-1].astype(bool)
Now lets create an empty segmentation map and occupancy map:
seg_map = np.zeros_like(shorted_masks[0],dtype=np.uint8)
occupancy_mask = np.zeros_like(shorted_masks[0],dtype=bool)
Next, we add the masks one by one (from high to low score) to the segmentation map. We only add a mask if it’s consistent with the masks that were previously added, which means only if the mask we want to add has less than 15% overlap with already occupied areas.
for i in range(shorted_masks.shape[0]):
mask = shorted_masks[i]
if (mask*occupancy_mask).sum()/mask.sum()>0.15: continue
mask[occupancy_mask]=0
seg_map[mask]=i+1
occupancy_mask[mask]=1
And this is it.
seg_mask now contains the predicted segmentation map with different values for each segment and 0 for the background.
We can turn this into a color map using:
rgb_image = np.zeros((seg_map.shape[0], seg_map.shape[1], 3), dtype=np.uint8)
for id_class in range(1,seg_map.max()+1):
rgb_image[seg_map == id_class] = [np.random.randint(255), np.random.randint(255), np.random.randint(255)]
And display:
cv2.imshow("annotation",rgb_image)
cv2.imshow("mix",(rgb_image/2+image/2).astype(np.uint8))
cv2.imshow("image",image)
cv2.waitKey()

The full inference code is available at:
Conclusion:
That’s it, we have trained and tested SAM2 on a custom dataset. Other than changing the data-reader, this should work for any dataset. In many cases, this should be enough to give a significant improvement in performance.
If this is not the case, there is more we can do: Since we only fine-tuned the final part of the net (mask-decoder) we gave it a limited capacity to learn. The main part of SAM2 is the image encoder. This is the bulk part of the net and fine-tuning it will take more data and a stronger GPU, but will give the net more room for improvement.
You can train this part by adding the command:
predictor.model.image_encoder.train(True)
Note that in this case, you will also need to scan the SAM2 code for “no_grad” commands and remove them (“ no_grad” blocks the gradient collection, which saves memory but prevents training).
Finally, SAM2 can also segment and track objects in videos, but fine-tuning this part is for another time.
Copyright: All images for the post are taken from the SAM2 GIT repository (under Apache license), and LabPics dataset (under MIT license). This tutorial code and nets are available under the Apache license.
Train/Fine-Tune Segment Anything 2 (SAM 2) in 60 Lines of Code was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
Originally appeared here:
Train/Fine-Tune Segment Anything 2 (SAM 2) in 60 Lines of Code
Go Here to Read this Fast! Train/Fine-Tune Segment Anything 2 (SAM 2) in 60 Lines of Code