Be mindful of the measure you choose
Testing and benchmarking machine learning models by comparing their predictions on a test set, even after deployment, is of fundamental importance. To do this, one needs to think of a measure or score that takes a prediction and a test point and assigns a value measuring how successful the prediction is with respect to the test point. However, one should think carefully about which scoring measure is appropriate. In particular, when choosing a method to evaluate a prediction we should adhere to the idea of proper scoring rules. I only give a loose definition of this idea here, but basically, we want a score that is minimized at the thing we want to measure!
As a general rule: One can use MSE to evaluate mean predictions, MAE to evaluate median predictions, the quantile score to evaluate more general quantile predictions and the energy or MMD score to evaluate distributional predictions.
Consider a variable you want to predict, say a random variable Y, from a vector of covariates X. In the example below, Y will be income and X will be certain characteristics, such as age and education. We learned a predictor f on some training data and now we predict Y as f(x). Usually, when we want to predict a variable Y as well as possible we predict the expectation of y given x, i.e. f(x) should approximate E[Y | X=x]. But more generally, f(x) could be an estimator of the median, other quantiles, or even the full conditional distribution P(Y | X=x).
Now for a new test point y, we want to score your prediction, that is you want a function S(y,f(x)), that is minimized (in expectation) when f(x) is the best thing you can do. For instance, if we want to predict E[Y | X=x], this score is given as the MSE: S(y, f(x))= (y-f(x))².
Here we study the principle of scoring the predictor f over at test set of (y_i,x_i), i=1,…,ntest in more detail. In all examples we will compare the ideal estimation method to an other that is clearly wrong, or naive, and show that our scores do what they are supposed to.
The Example
To illustrate things, I will simulate a simple dataset that should mimic income data. We will use this simple example throughout this article to illustrate the concepts.
library(dplyr)
#Create some variables:
# Simulate data for 100 individuals
n <- 5000
# Generate age between 20 and 60
age <- round(runif(n, min = 20, max = 60))
# Define education levels
education_levels <- c("High School", "Bachelor's", "Master's")
# Simulate education level probabilities
education_probs <- c(0.4, 0.4, 0.2)
# Sample education level based on probabilities
education <- sample(education_levels, n, replace = TRUE, prob = education_probs)
# Simulate experience correlated with age with some random error
experience <- age - 20 + round(rnorm(n, mean = 0, sd = 3))
# Define a non-linear function for wage
wage <- exp((age * 0.1) + (case_when(education == "High School" ~ 1,
education == "Bachelor's" ~ 1.5,
TRUE ~ 2)) + (experience * 0.05) + rnorm(n, mean = 0, sd = 0.5))
hist(wage)
Although this simulation may be oversimplified, it reflects certain well-known characteristics of such data: older age, advanced education, and greater experience are all linked to higher wages. The use of the “exp” operator results in a highly skewed wage distribution, which is a consistent observation in such datasets.

Crucially, this skewness is also present when we fix age, education and experience to certain values. Let’s imagine we look at a specific person, Dave, who is 30 years old, has a Bachelor’s in Economics and 10 years of experience and let’s look at his actual income distribution according to our data generating process:
ageDave<-30
educationDave<-"Bachelor's"
experienceDave <- 10
wageDave <- exp((ageDave * 0.1) + (case_when(educationDave == "High School" ~ 1,
educationDave == "Bachelor's" ~ 1.5,
TRUE ~ 2)) + (experienceDave * 0.05) + rnorm(n, mean = 0, sd = 0.5))
hist(wageDave, main="Wage Distribution for Dave", xlab="Wage")

Thus the distribution of possible wages of Dave, given the information we have about him, is still highly skewed.
We also generate a test set of several people:
## Generate test set
ntest<-1000
# Generate age between 20 and 60
agetest <- round(runif(ntest, min = 20, max = 60))
# Sample education level based on probabilities
educationtest <- sample(education_levels, ntest, replace = TRUE, prob = education_probs)
# Simulate experience correlated with age with some random error
experiencetest <- agetest - 20 + round(rnorm(ntest, mean = 0, sd = 3))
## Generate ytest that we try to predict:
wagetest <- exp((agetest * 0.1) + (case_when(educationtest == "High School" ~ 1,
educationtest == "Bachelor's" ~ 1.5,
TRUE ~ 2)) + (experiencetest * 0.05) + rnorm(ntest, mean = 0, sd = 0.5))
We now start simple and first look at the scores for mean and median prediction.
The scores for mean and median prediction
In data science and machine learning, interest often centers on a single number that signifies the “center” or “middle” of the distribution we aim to predict, namely the (conditional) mean or median. To do this we have the mean squared error (MSE):

and the mean absolute error (MAE):

An important takeaway is that the MSE is the appropriate metric for predicting the conditional mean, while the MAE is the measure to use for the conditional median. Mean and median are not the same thing for skewed distributions like the one we study here.
Let us illustrate this for the above example with very simple estimators (that we would not have access to in real life), just for illustration:
conditionalmeanest <-
function(age, education, experience, N = 1000) {
mean(exp((age * 0.1) + (
case_when(
education == "High School" ~ 1,
education == "Bachelor's" ~ 1.5,
TRUE ~ 2
)
) + (experience * 0.05) + rnorm(N, mean = 0, sd = 0.5)
))
}
conditionalmedianest <-
function(age, education, experience, N = 1000) {
median(exp((age * 0.1) + (
case_when(
education == "High School" ~ 1,
education == "Bachelor's" ~ 1.5,
TRUE ~ 2
)
) + (experience * 0.05) + rnorm(N, mean = 0, sd = 0.5)
))
}
That is we estimate mean and median, by simply simulating from the model for fixed values of age, education, and experience (this would be a simulation from the correct conditional distribution) and then we simply take the mean/median of that. Let’s test this on Dave:
hist(wageDave, main="Wage Distribution for Dave", xlab="Wage")
abline(v=conditionalmeanest(ageDave, educationDave, experienceDave), col="darkred", cex=1.2)
abline(v=conditionalmedianest(ageDave, educationDave, experienceDave), col="darkblue", cex=1.2)

Clearly the mean and median are different, as one would expect from such a distribution. In fact, as is typical for income distributions, the mean is higher (more influenced by high values) than the median.
Now let’s use these estimators on the test set:
Xtest<-data.frame(age=agetest, education=educationtest, experience=experiencetest)
meanest<-sapply(1:nrow(Xtest), function(j) conditionalmeanest(Xtest$age[j], Xtest$education[j], Xtest$experience[j]) )
median<-sapply(1:nrow(Xtest), function(j) conditionalmedianest(Xtest$age[j], Xtest$education[j], Xtest$experience[j]) )
This gives a diverse range of conditional mean/median values. Now we calculate MSE and MAE:
(MSE1<-mean((meanest-wagetest)^2))
(MSE2<-mean((median-wagetest)^2))
MSE1 < MSE2
### Method 1 (the true mean estimator) is better than method 2!
# but the MAE is actually worse of method 1!
(MAE1<-mean(abs(meanest-wagetest)) )
(MAE2<-mean( abs(median-wagetest)))
MAE1 < MAE2
### Method 2 (the true median estimator) is better than method 1!
This shows what is known theoretically: MSE is minimized for the (conditional) expectation E[Y | X=x], while MAE is minimized at the conditional median. In general, it does not make sense to use the MAE when you try to evaluate your mean prediction. In a lot of applied research and data science, people use the MAE or both to evaluate mean predictions (I know because I did it myself). While this may be warranted in certain applications, this can have serious consequences for distributions that are not symmetric, as we saw in this example: When looking at the MAE, method 1 looks worse than method 2, even though the former estimates the mean correctly. In fact, in this highly skewed example, method 1 should have a lower MAE than method 2.
To score conditional mean prediction use the mean squared error (MSE) and not the mean absolute error (MAE). The MAE is minimized for the conditional median.
Scores for quantile and interval prediction
Assume we want to score an estimate f(x) of the quantile q_x such that
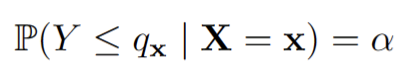

In this case, we can consider the quantile score:

whereby

To unpack this formula, we can consider two cases:
(1) y is smaller than f(x):
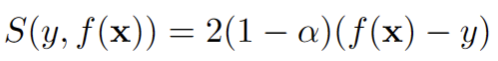
i.e. we incur a penalty which gets bigger the further away y is from f(x).
(2) y is larger than f(x):

i.e. a penalty which gets bigger the further away y is from f(x).
Notice that the weight is such that for a high alpha, having the estimated quantile f(x) smaller than y gets penalized more. This is by design and ensures that the right quantile is indeed the minimizer of the expected value of S(y,f(x)) over y. This score is in fact the quantile loss (up to a factor 2), see e.g. this nice article. It is implemented in the quantile_score function of the package scoringutils in R. Finally, note that for alpha=0.5:

simply the MAE! This makes sense, as the 0.5 quantile is the median.
With the power to predict quantiles, we can also build prediction intervals. Consider (l_x, u_x), where l_x ≤ u_x are quantiles such that

In fact, this is met if l_x is the alpha/2 quantile, and u_x is the 1-alpha/2 quantile. Thus we now estimate and score these two quantiles. Consider f(x)=(f_1(x), f_2(x)), whereby f_1(x) to be an estimate of l_x and f_2(x) an estimate of u_x. We provide two estimators, the “ideal” one that simulates again from the true process to then estimate the required quantiles and a “naive” one, which has the right coverage but is too big:
library(scoringutils)
## Define conditional quantile estimation
conditionalquantileest <-
function(probs, age, education, experience, N = 1000) {
quantile(exp((age * 0.1) + (
case_when(
education == "High School" ~ 1,
education == "Bachelor's" ~ 1.5,
TRUE ~ 2
)
) + (experience * 0.05) + rnorm(N, mean = 0, sd = 0.5)
)
, probs =
probs)
}
## Define a very naive estimator that will still have the required coverage
lowernaive <- 0
uppernaive <- max(wage)
# Define the quantile of interest
alpha <- 0.05
lower <-
sapply(1:nrow(Xtest), function(j)
conditionalquantileest(alpha / 2, Xtest$age[j], Xtest$education[j], Xtest$experience[j]))
upper <-
sapply(1:nrow(Xtest), function(j)
conditionalquantileest(1 - alpha / 2, Xtest$age[j], Xtest$education[j], Xtest$experience[j]))
## Calculate the scores for both estimators
# 1. Score the alpha/2 quantile estimate
qs_lower <- mean(quantile_score(wagetest,
predictions = lower,
quantiles = alpha / 2))
# 2. Score the alpha/2 quantile estimate
qs_upper <- mean(quantile_score(wagetest,
predictions = upper,
quantiles = 1 - alpha / 2))
# 1. Score the alpha/2 quantile estimate
qs_lowernaive <- mean(quantile_score(wagetest,
predictions = rep(lowernaive, ntest),
quantiles = alpha / 2))
# 2. Score the alpha/2 quantile estimate
qs_uppernaive <- mean(quantile_score(wagetest,
predictions = rep(uppernaive, ntest),
quantiles = 1 - alpha / 2))
# Construct the interval score by taking the average
(interval_score <- (qs_lower + qs_upper) / 2)
# Score of the ideal estimator: 187.8337
# Construct the interval score by taking the average
(interval_scorenaive <- (qs_lowernaive + qs_uppernaive) / 2)
# Score of the naive estimator: 1451.464
Again we can clearly see that, on average, the correct estimator has a much lower score than the naive one!
Thus with the quantile score, we have a reliable way of scoring individual quantile predictions. However, the way of averaging the score of the upper and lower quantiles for the prediction interval might seem ad hoc. Luckily it turns out that this leads to the so-called interval score:
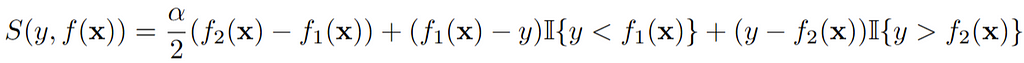
Thus through some algebraic magic, we can score a prediction interval by averaging the scores for the alpha/2 and the 1-alpha/2 quantiles as we did. Interestingly, the resulting interval score rewards narrow prediction intervals, and induces a penalty, the size of which depends on alpha, if the observation misses the interval. Instead of using the average of quantile scores, we can also directly calculate this score with the package scoringutils.
alpha <- 0.05
mean(interval_score(
wagetest,
lower=lower,
upper=upper,
interval_range=(1-alpha)*100,
weigh = T,
separate_results = FALSE
))
#Score of the ideal estimator: 187.8337
This is the exact same number we got above when averaging the scores of the two intervals.
The quantile score implemented in R in the package scoringutils can be used to score quantile predictions. If one wants to score a prediction interval directly, the interval_score function can be used.
Scores for distributional prediction
More and more fields have to deal with distributional prediction. Luckily there are even scores for this problem. In particular, here I focus on what is called the energy score:

for f(x) being an estimate of the distribution P(Y | X=x). The second term takes the expectation of the Eucledian distance between two independent samples from f(x). This is akin to a normalizing term, establishing the value if the same distribution was compared. The first term then compares the sample point y to a draw X from f(x). In expectation (over Y drawn from P(Y | X=x)) this will be minimized if f(x)=P(Y | X=x).
Thus instead of just predicting the mean or the quantiles, we now try to predict the whole distribution of wage at each test point. Essentially we try to predict and evaluate the conditional distribution we plotted for Dave above. This is a bit more complicated; how exactly do we represent a learned distribution? In practice this is resolved by assuming we can obtain a sample from the predicted distribution. Thus we compare a sample of N, obtained from the predicted distribution, to a single test point. This can be done in R using es_sample from the scoringRules package:
library(scoringRules)
## Ideal "estimate": Simply sample from the true conditional distribution
## P(Y | X=x) for each sample point x
distributionestimate <-
function(age, education, experience, N = 100) {
exp((age * 0.1) + (
case_when(
education == "High School" ~ 1,
education == "Bachelor's" ~ 1.5,
TRUE ~ 2
)
) + (experience * 0.05) + rnorm(N, mean = 0, sd = 0.5))
}
## Naive Estimate: Only sample from the error distribution, without including the
## information of each person.
distributionestimatenaive <-
function(age, education, experience, N = 100) {
exp(rnorm(N, mean = 0, sd = 0.5))
}
scoretrue <- mean(sapply(1:nrow(Xtest), function(j) {
wageest <-
distributionestimate(Xtest$age[j], Xtest$education[j], Xtest$experience[j])
return(scoringRules::es_sample(y = wagetest[j], dat = matrix(wageest, nrow=1)))
}))
scorenaive <- mean(sapply(1:nrow(Xtest), function(j) {
wageest <-
distributionestimatenaive(Xtest$age[j], Xtest$education[j], Xtest$experience[j])
return(scoringRules::es_sample(y = wagetest[j], dat = matrix(wageest, nrow=1)))
}))
## scoretrue: 761.026
## scorenaive: 2624.713
In the above code, we again compare the “perfect” estimate (i.e. sampling from the true distribution P(Y | X=x)) to a very naive one, namely one that does not consider any information on wage, edicuation or experience. Again, the score reliably identifies the better of the two methods.
The energy score, implemented in the R package scoringRules can be used to score distributional prediction, if a sample from the predicted distribution is available.
Conclusion
We have looked at different ways of scoring predictions. Thinking about the right measure to test predictions is important, as the wrong measure might make us choose and keep the wrong model for our prediction task.
It should be noted that especially for distributional prediction this scoring is a difficult task and the score might not have much power in practice. That is, even a method that leads to a large improvement might only have a slightly smaller score. However, this is not a problem per se, as long as the score is able to reliably identify the better of the two methods.
References
[1] Tilmann Gneiting & Adrian E Raftery (2007) Strictly Proper Scoring Rules, Prediction, and Estimation, Journal of the American Statistical Association, 102:477, 359–378, DOI: 10.1198/016214506000001437
Appendix: All the code in one place
library(dplyr)
#Create some variables:
# Simulate data for 100 individuals
n <- 5000
# Generate age between 20 and 60
age <- round(runif(n, min = 20, max = 60))
# Define education levels
education_levels <- c("High School", "Bachelor's", "Master's")
# Simulate education level probabilities
education_probs <- c(0.4, 0.4, 0.2)
# Sample education level based on probabilities
education <- sample(education_levels, n, replace = TRUE, prob = education_probs)
# Simulate experience correlated with age with some random error
experience <- age - 20 + round(rnorm(n, mean = 0, sd = 3))
# Define a non-linear function for wage
wage <- exp((age * 0.1) + (case_when(education == "High School" ~ 1,
education == "Bachelor's" ~ 1.5,
TRUE ~ 2)) + (experience * 0.05) + rnorm(n, mean = 0, sd = 0.5))
hist(wage)
ageDave<-30
educationDave<-"Bachelor's"
experienceDave <- 10
wageDave <- exp((ageDave * 0.1) + (case_when(educationDave == "High School" ~ 1,
educationDave == "Bachelor's" ~ 1.5,
TRUE ~ 2)) + (experienceDave * 0.05) + rnorm(n, mean = 0, sd = 0.5))
hist(wageDave, main="Wage Distribution for Dave", xlab="Wage")
## Generate test set
ntest<-1000
# Generate age between 20 and 60
agetest <- round(runif(ntest, min = 20, max = 60))
# Sample education level based on probabilities
educationtest <- sample(education_levels, ntest, replace = TRUE, prob = education_probs)
# Simulate experience correlated with age with some random error
experiencetest <- agetest - 20 + round(rnorm(ntest, mean = 0, sd = 3))
## Generate ytest that we try to predict:
wagetest <- exp((agetest * 0.1) + (case_when(educationtest == "High School" ~ 1,
educationtest == "Bachelor's" ~ 1.5,
TRUE ~ 2)) + (experiencetest * 0.05) + rnorm(ntest, mean = 0, sd = 0.5))
conditionalmeanest <-
function(age, education, experience, N = 1000) {
mean(exp((age * 0.1) + (
case_when(
education == "High School" ~ 1,
education == "Bachelor's" ~ 1.5,
TRUE ~ 2
)
) + (experience * 0.05) + rnorm(N, mean = 0, sd = 0.5)
))
}
conditionalmedianest <-
function(age, education, experience, N = 1000) {
median(exp((age * 0.1) + (
case_when(
education == "High School" ~ 1,
education == "Bachelor's" ~ 1.5,
TRUE ~ 2
)
) + (experience * 0.05) + rnorm(N, mean = 0, sd = 0.5)
))
}
hist(wageDave, main="Wage Distribution for Dave", xlab="Wage")
abline(v=conditionalmeanest(ageDave, educationDave, experienceDave), col="darkred", cex=1.2)
abline(v=conditionalmedianest(ageDave, educationDave, experienceDave), col="darkblue", cex=1.2)
Xtest<-data.frame(age=agetest, education=educationtest, experience=experiencetest)
meanest<-sapply(1:nrow(Xtest), function(j) conditionalmeanest(Xtest$age[j], Xtest$education[j], Xtest$experience[j]) )
median<-sapply(1:nrow(Xtest), function(j) conditionalmedianest(Xtest$age[j], Xtest$education[j], Xtest$experience[j]) )
(MSE1<-mean((meanest-wagetest)^2))
(MSE2<-mean((median-wagetest)^2))
MSE1 < MSE2
### Method 1 (the true mean estimator) is better than method 2!
# but the MAE is actually worse of method 1!
(MAE1<-mean(abs(meanest-wagetest)) )
(MAE2<-mean( abs(median-wagetest)))
MAE1 < MAE2
### Method 2 (the true median estimator) is better than method 1!
library(scoringutils)
## Define conditional quantile estimation
conditionalquantileest <-
function(probs, age, education, experience, N = 1000) {
quantile(exp((age * 0.1) + (
case_when(
education == "High School" ~ 1,
education == "Bachelor's" ~ 1.5,
TRUE ~ 2
)
) + (experience * 0.05) + rnorm(N, mean = 0, sd = 0.5)
)
, probs =
probs)
}
## Define a very naive estimator that will still have the required coverage
lowernaive <- 0
uppernaive <- max(wage)
# Define the quantile of interest
alpha <- 0.05
lower <-
sapply(1:nrow(Xtest), function(j)
conditionalquantileest(alpha / 2, Xtest$age[j], Xtest$education[j], Xtest$experience[j]))
upper <-
sapply(1:nrow(Xtest), function(j)
conditionalquantileest(1 - alpha / 2, Xtest$age[j], Xtest$education[j], Xtest$experience[j]))
## Calculate the scores for both estimators
# 1. Score the alpha/2 quantile estimate
qs_lower <- mean(quantile_score(wagetest,
predictions = lower,
quantiles = alpha / 2))
# 2. Score the alpha/2 quantile estimate
qs_upper <- mean(quantile_score(wagetest,
predictions = upper,
quantiles = 1 - alpha / 2))
# 1. Score the alpha/2 quantile estimate
qs_lowernaive <- mean(quantile_score(wagetest,
predictions = rep(lowernaive, ntest),
quantiles = alpha / 2))
# 2. Score the alpha/2 quantile estimate
qs_uppernaive <- mean(quantile_score(wagetest,
predictions = rep(uppernaive, ntest),
quantiles = 1 - alpha / 2))
# Construct the interval score by taking the average
(interval_score <- (qs_lower + qs_upper) / 2)
# Score of the ideal estimator: 187.8337
# Construct the interval score by taking the average
(interval_scorenaive <- (qs_lowernaive + qs_uppernaive) / 2)
# Score of the naive estimator: 1451.464
library(scoringRules)
## Ideal "estimate": Simply sample from the true conditional distribution
## P(Y | X=x) for each sample point x
distributionestimate <-
function(age, education, experience, N = 100) {
exp((age * 0.1) + (
case_when(
education == "High School" ~ 1,
education == "Bachelor's" ~ 1.5,
TRUE ~ 2
)
) + (experience * 0.05) + rnorm(N, mean = 0, sd = 0.5))
}
## Naive Estimate: Only sample from the error distribution, without including the
## information of each person.
distributionestimatenaive <-
function(age, education, experience, N = 100) {
exp(rnorm(N, mean = 0, sd = 0.5))
}
scoretrue <- mean(sapply(1:nrow(Xtest), function(j) {
wageest <-
distributionestimate(Xtest$age[j], Xtest$education[j], Xtest$experience[j])
return(scoringRules::es_sample(y = wagetest[j], dat = matrix(wageest, nrow=1)))
}))
scorenaive <- mean(sapply(1:nrow(Xtest), function(j) {
wageest <-
distributionestimatenaive(Xtest$age[j], Xtest$education[j], Xtest$experience[j])
return(scoringRules::es_sample(y = wagetest[j], dat = matrix(wageest, nrow=1)))
}))
## scoretrue: 761.026
## scorenaive: 2624.713
How to Evaluate Your Predictions was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
Originally appeared here:
How to Evaluate Your Predictions