
Read and write dataframes up to ten times faster than Parquet with StaticFrame NPZ

The Apache Parquet format provides an efficient binary representation of columnar table data, as seen with widespread use in Apache Hadoop and Spark, AWS Athena and Glue, and Pandas DataFrame serialization. While Parquet offers broad interoperability with performance superior to text formats (such as CSV or JSON), it is as much as ten times slower than NPZ, an alternative DataFrame serialization format introduced in StaticFrame.
StaticFrame (an open-source DataFrame library of which I am an author) builds upon NumPy NPY and NPZ formats to encode DataFrames. The NPY format (a binary encoding of array data) and the NPZ format (zipped bundles of NPY files) are defined in a NumPy Enhancement Proposal from 2007. By extending the NPZ format with specialized JSON metadata, StaticFrame provides a complete DataFrame serialization format that supports all NumPy dtypes.
This article extends work first presented at PyCon USA 2022 with further performance optimizations and broader benchmarking.
The Challenge of Serializing DataFrames
DataFrames are not just collections of columnar data with string column labels, such as found in relational databases. In addition to columnar data, DataFrames have labelled rows and columns, and those row and column labels can be of any type or (with hierarchical labels) many types. Further, it is common to store metadata with a name attribute, either on the DataFrame or on the axis labels.
As Parquet was originally designed just to store collections of columnar data, the full range of DataFrame characteristics is not directly supported. Pandas supplies this additional information by adding JSON metadata into the Parquet file.
Further, Parquet supports a minimal selection of types; the full range of NumPy dtypes is not directly supported. For example, Parquet does not natively support unsigned integers or any date types.
While Python pickles are capable of efficiently serializing DataFrames and NumPy arrays, they are only suitable for short-term caches from trusted sources. While pickles are fast, they can become invalid due to code changes and are insecure to load from untrusted sources.
Another alternative to Parquet, originating in the Arrow project, is Feather. While Feather supports all Arrow types and succeeds in being faster than Parquet, it is still at least two times slower reading DataFrames than NPZ.
Parquet and Feather support compression to reduce file size. Parquet defaults to using “snappy” compression, while Feather defaults to “lz4”. As the NPZ format prioritizes performance, it does not yet support compression. As will be shown below, NPZ outperforms both compressed and uncompressed Parquet files by significant factors.
DataFrame Serialization Performance Comparisons
Numerous publications offer DataFrame benchmarks by testing just one or two datasets. McKinney and Richardson (2020) is an example, where two datasets, Fannie Mae Loan Performance and NYC Yellow Taxi Trip data, are used to generalize about performance. Such idiosyncratic datasets are insufficient, as both the shape of the DataFrame and the degree of columnar type heterogeneity can significantly differentiate performance.
To avoid this deficiency, I compare performance with a panel of nine synthetic datasets. These datasets vary along two dimensions: shape (tall, square, and wide) and columnar heterogeneity (columnar, mixed, and uniform). Shape variations alter the distribution of elements between tall (e.g., 10,000 rows and 100 columns), square (e.g., 1,000 rows and columns), and wide (e.g., 100 rows and 10,000 columns) geometries. Columnar heterogeneity variations alter the diversity of types between columnar (no adjacent columns have the same type), mixed (some adjacent columns have the same type), and uniform (all columns have the same type).
The frame-fixtures library defines a domain-specific language to create deterministic, randomly-generated DataFrames for testing; the nine datasets are generated with this tool.
To demonstrate some of the StaticFrame and Pandas interfaces evaluated, the following IPython session performs basic performance tests using %time. As shown below, a square, uniformly-typed DataFrame can be written and read with NPZ many times faster than uncompressed Parquet.
>>> import numpy as np
>>> import static_frame as sf
>>> import pandas as pd
>>> # an square, uniform float array
>>> array = np.random.random_sample((10_000, 10_000))
>>> # write peformance
>>> f1 = sf.Frame(array)
>>> %time f1.to_npz('/tmp/frame.npz')
CPU times: user 710 ms, sys: 396 ms, total: 1.11 s
Wall time: 1.11 s
>>> df1 = pd.DataFrame(array)
>>> %time df1.to_parquet('/tmp/df.parquet', compression=None)
CPU times: user 6.82 s, sys: 900 ms, total: 7.72 s
Wall time: 7.74 s
>>> # read performance
>>> %time f2 = f1.from_npz('/tmp/frame.npz')
CPU times: user 2.77 ms, sys: 163 ms, total: 166 ms
Wall time: 165 ms
>>> %time df2 = pd.read_parquet('/tmp/df.parquet')
CPU times: user 2.55 s, sys: 1.2 s, total: 3.75 s
Wall time: 866 ms
Performance tests provided below extend this basic approach by using frame-fixtures for systematic variation of shape and type heterogeneity, and average results over ten iterations. While hardware configuration will affect performance, relative characteristics are retained across diverse machines and operating systems. For all interfaces the default parameters are used, except for disabling compression as needed. The code used to perform these tests is available at GitHub.
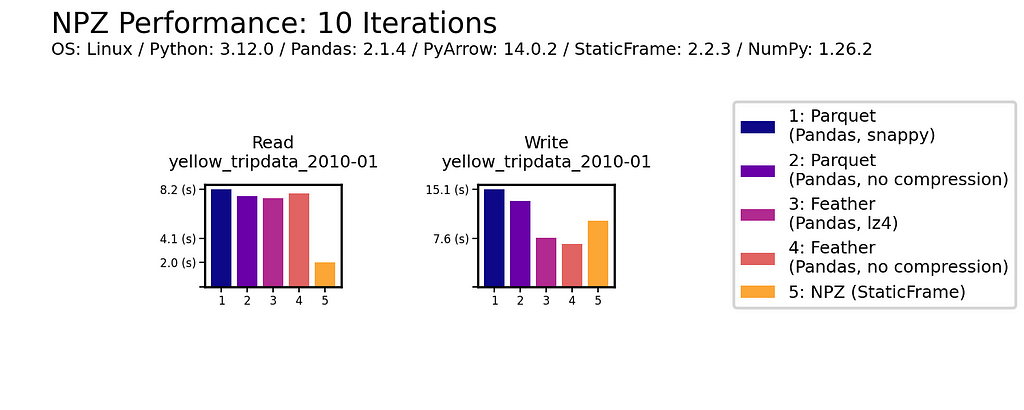
Read Performance
As data is generally read more often then it is written, read performance is a priority. As shown for all nine DataFrames of one million (1e+06) elements, NPZ significantly outperforms Parquet and Feather with every fixture. NPZ read performance is over ten times faster than compressed Parquet. For example, with the Uniform Tall fixture, compressed Parquet reading is 21 ms compared to 1.5 ms with NPZ.
The chart below shows processing time, where lower bars correspond to faster performance.
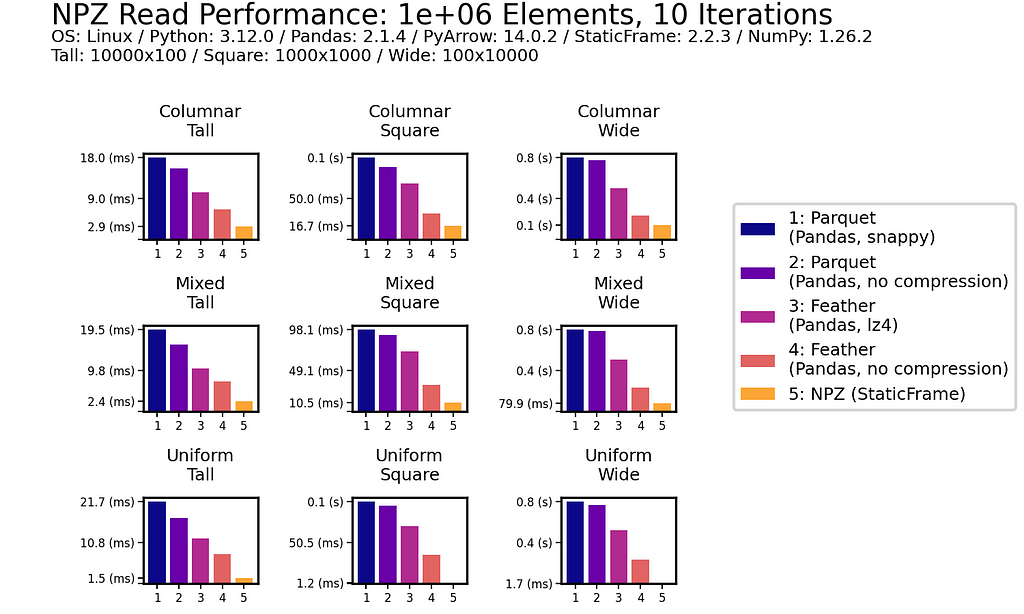
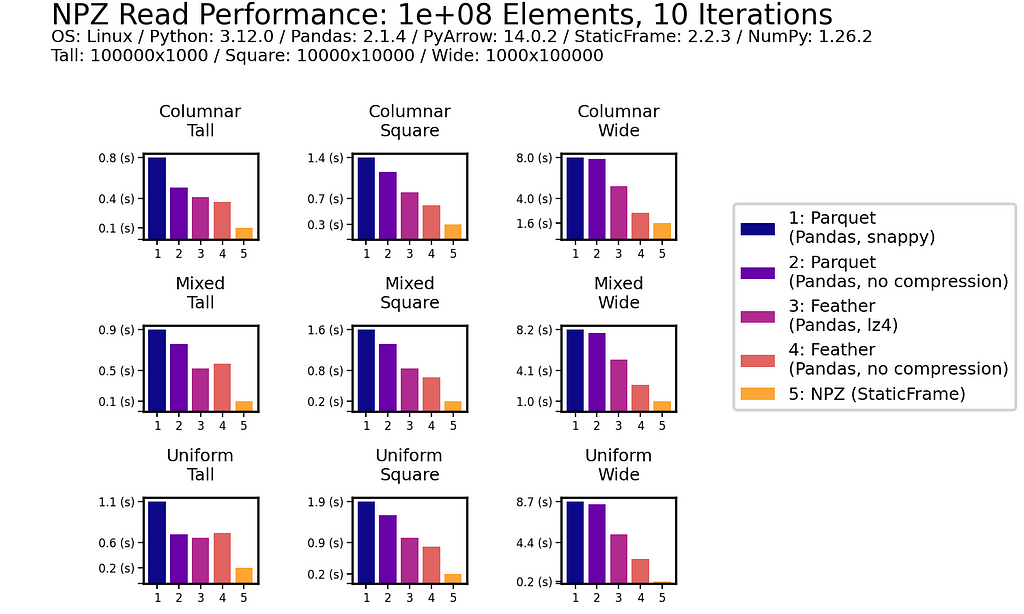
This impressive NPZ performance is retained with scale. Moving to 100 million (1e+08) elements, NPZ continues to perform at least twice as fast as Parquet and Feather, regardless of if compression is used.
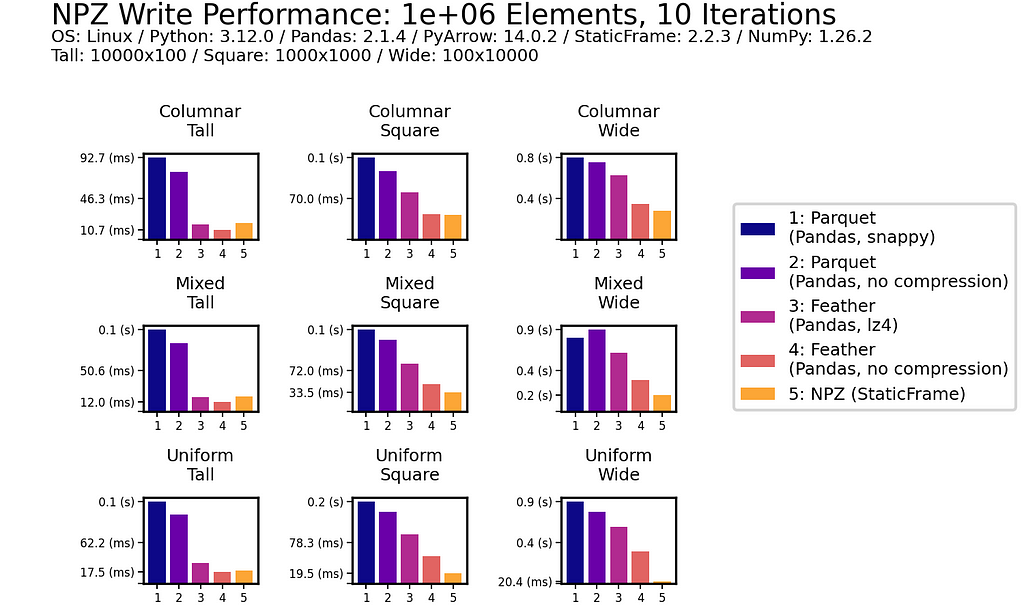
Write Performance
In writing DataFrames to disk, NPZ outperforms Parquet (both compressed and uncompressed) in all scenarios. For example, with the Uniform Square fixture, compressed Parquet writing is 200 ms compared to 18.3 ms with NPZ. NPZ write performance is generally comparable to uncompressed Feather: in some scenarios NPZ is faster, in others, Feather is faster.
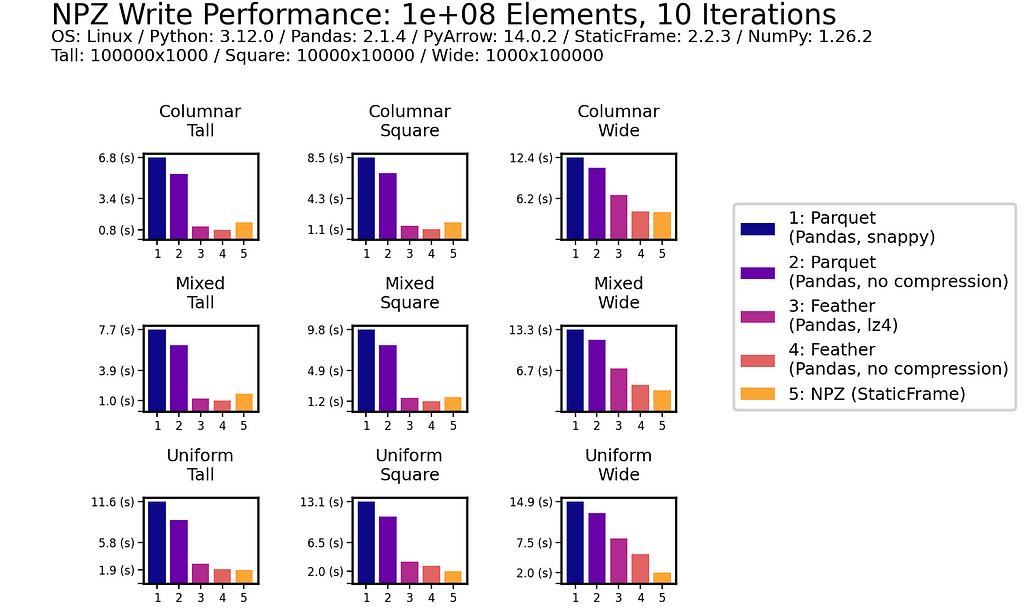
As with read performance, NPZ write performance is retained with scale. Moving to 100 million (1e+08) elements, NPZ continues to be at least twice as fast as Parquet, regardless of if compression is used or not.
Idiosyncratic Performance
As an additional reference, we will also benchmark the same NYC Yellow Taxi Trip data (from January 2010) used in McKinney and Richardson (2020). This dataset contains almost 300 million (3e+08) elements in a tall, heterogeneously typed DataFrame of 14,863,778 rows and 19 columns.
NPZ read performance is shown to be around four times faster than Parquet and Feather (with or without compression). While NPZ write performance is faster than Parquet, Feather writing here is fastest.
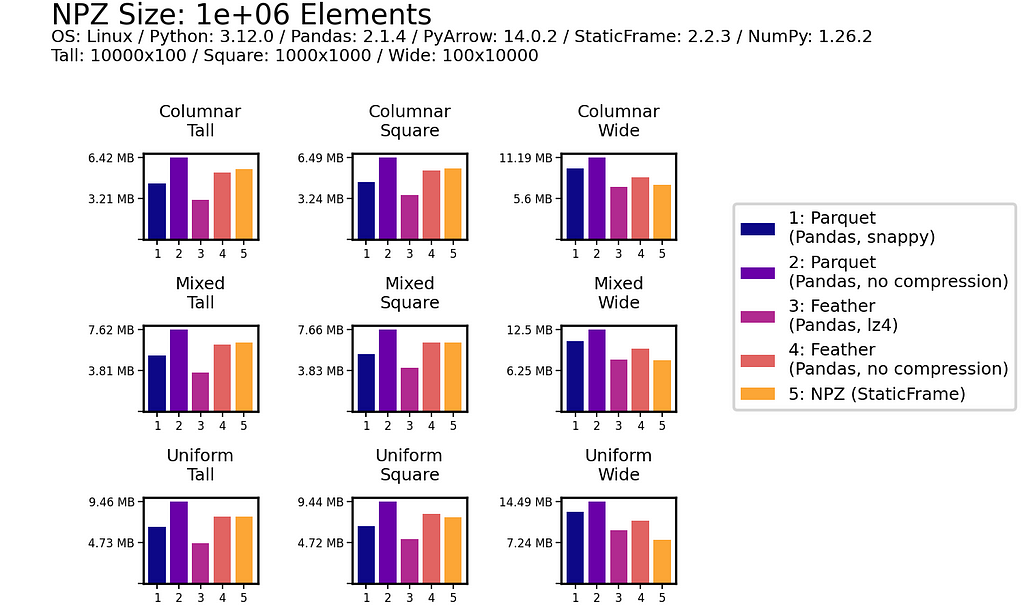
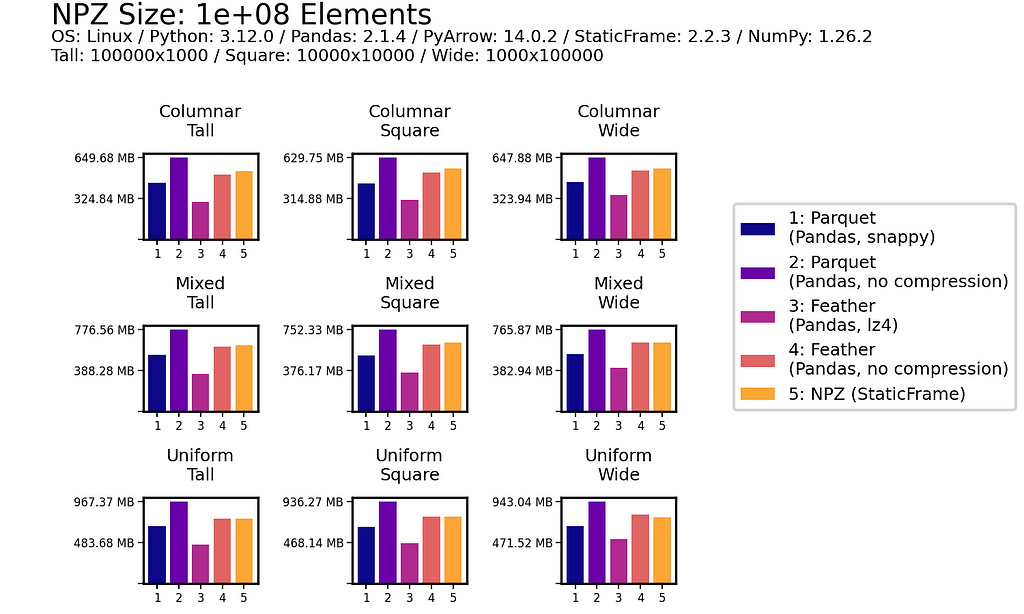
File Size
As shown below for one million (1e+06) element and 100 million (1e+08) element DataFrames, uncompressed NPZ is generally equal in size on disk to uncompressed Feather and always smaller than uncompressed Parquet (sometimes smaller than compressed Parquet too). As compression provides only modest file-size reductions for Parquet and Feather, the benefit of uncompressed NPZ in speed might easily outweigh the cost of greater size.
Serializing DataFrames
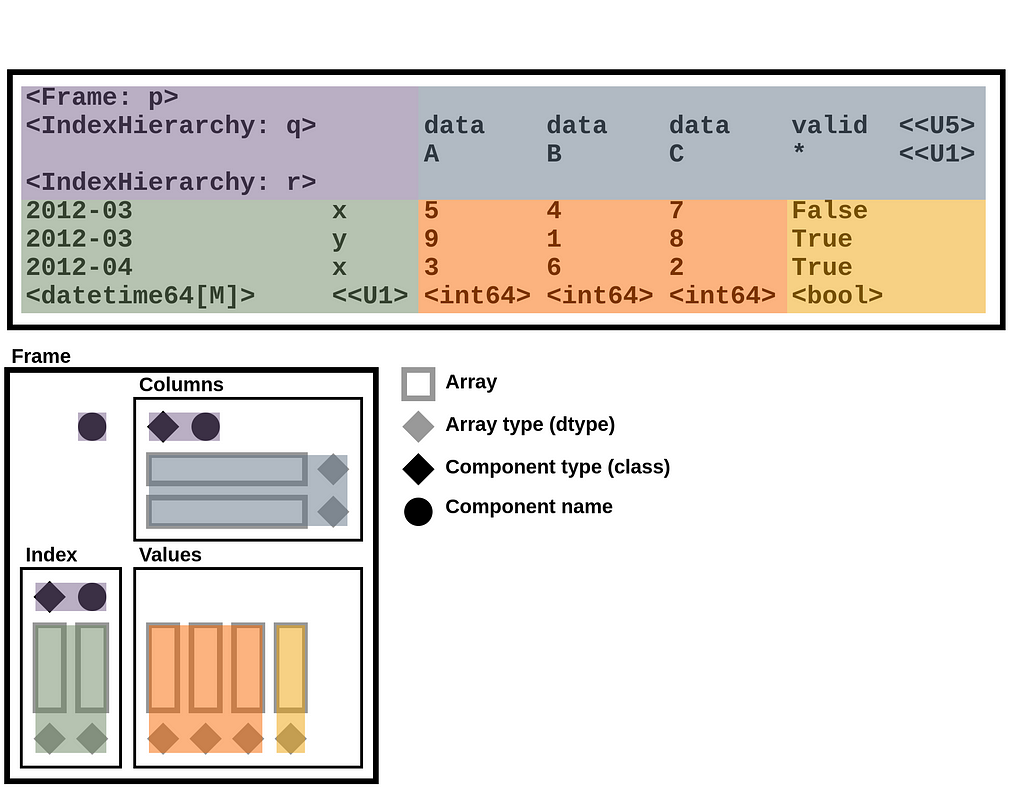
StaticFrame stores data as a collection of 1D and 2D NumPy arrays. Arrays represent columnar values, as well as variable-depth index and column labels. In addition to NumPy arrays, information about component types (i.e., the Python class used for the index and columns), as well as the component name attributes, are needed to fully reconstruct a Frame. Completely serializing a DataFrame requires writing and reading these components to a file.
DataFrame components can be represented by the following diagram, which isolates arrays, array types, component types, and component names. This diagram will be used to demonstrate how an NPZ encodes a DataFrame.
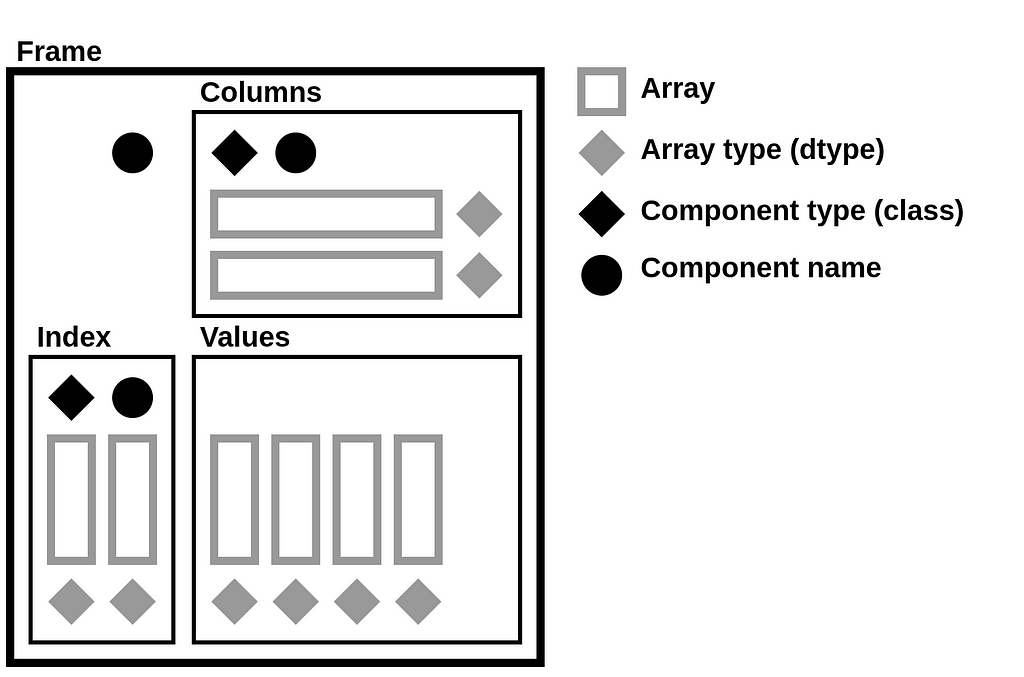
The components of that diagram map to components of a Frame string representation in Python. For example, given a Frame of integers and Booleans with hierarchical labels on both the index and columns (downloadable via GitHub with StaticFrame’s WWW interface), StaticFrame provides the following string representation:
>>> frame = sf.Frame.from_npz(sf.WWW.from_file('https://github.com/static-frame/static-frame/raw/master/doc/source/articles/serialize/frame.npz', encoding=None))
>>> frame
<Frame: p>
<IndexHierarchy: q> data data data valid <<U5>
A B C * <<U1>
<IndexHierarchy: r>
2012-03 x 5 4 7 False
2012-03 y 9 1 8 True
2012-04 x 3 6 2 True
<datetime64[M]> <<U1> <int64> <int64> <int64> <bool>
The components of the string representation can be mapped to the DataFrame diagram by color:
Encoding an Array in NPY
A NPY stores a NumPy array as a binary file with six components: (1) a “magic” prefix, (2) a version number, (3) a header length and (4) header (where the header is a string representation of a Python dictionary), and (5) padding followed by (6) raw array byte data. These components are shown below for a three-element binary array stored in a file named “__blocks_1__.npy”.
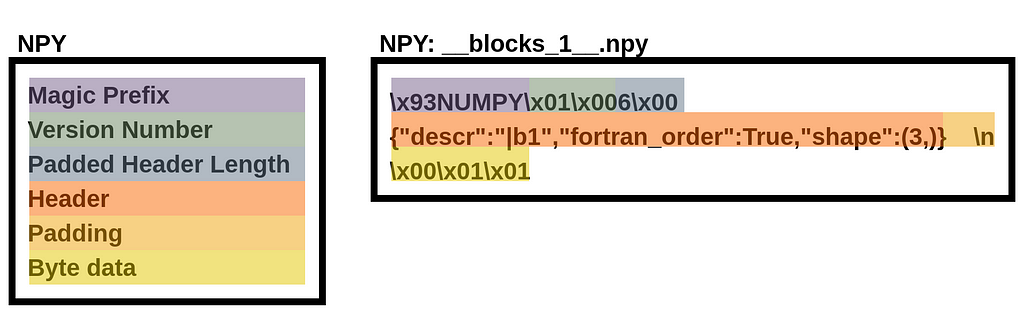
Given a NPZ file named “frame.npz”, we can extract the binary data by reading the NPY file from the NPZ with the standard library’s ZipFile:
>>> from zipfile import ZipFile
>>> with ZipFile('/tmp/frame.npz') as zf: print(zf.open('__blocks_1__.npy').read())
b'x93NUMPYx01x006x00{"descr":"|b1","fortran_order":True,"shape":(3,)} nx00x01x01
As NPY is well supported in NumPy, the np.load() function can be used to convert this file to a NumPy array. This means that underlying array data in a StaticFrame NPZ is easily extractable by alternative readers.
>>> with ZipFile('/tmp/frame.npz') as zf: print(repr(np.load(zf.open('__blocks_1__.npy'))))
array([False, True, True])
As a NPY file can encode any array, large two-dimensional arrays can be loaded from contiguous byte data, providing excellent performance in StaticFrame when multiple contiguous columns are represented by a single array.
Building a NPZ File
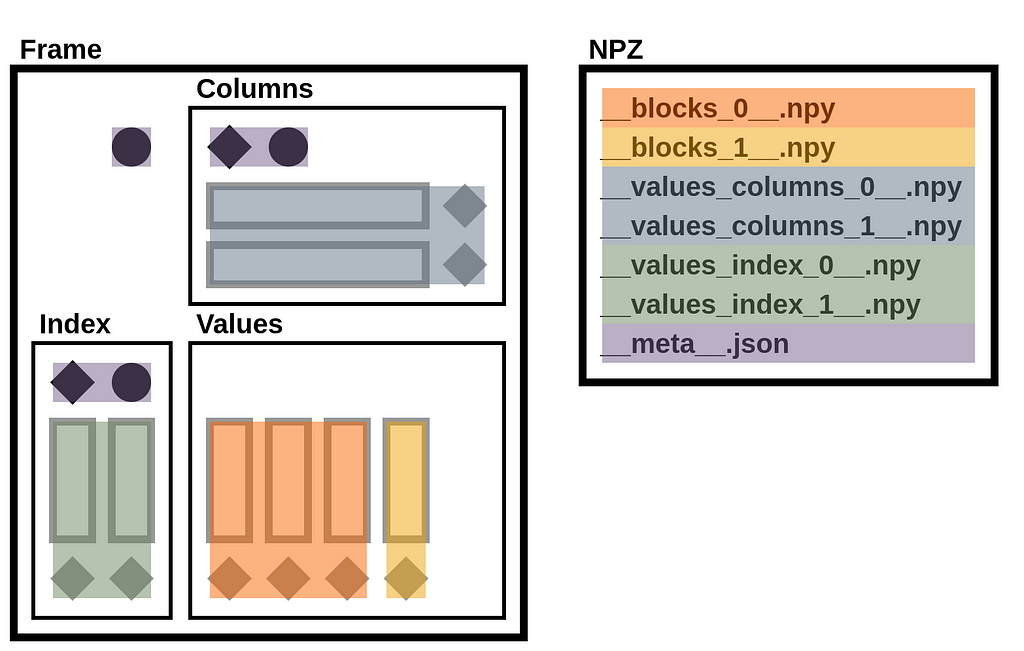
A StaticFrame NPZ is a standard uncompressed ZIP file that contains array data in NPY files and metadata (containing component types and names) in a JSON file.
Given the NPZ file for the Frame above, we can list its contents with ZipFile. The archive contains six NPY files and one JSON file.
>>> with ZipFile('/tmp/frame.npz') as zf: print(zf.namelist())
['__values_index_0__.npy', '__values_index_1__.npy', '__values_columns_0__.npy', '__values_columns_1__.npy', '__blocks_0__.npy', '__blocks_1__.npy', '__meta__.json']
The illustration below maps these files to components of the DataFrame diagram.
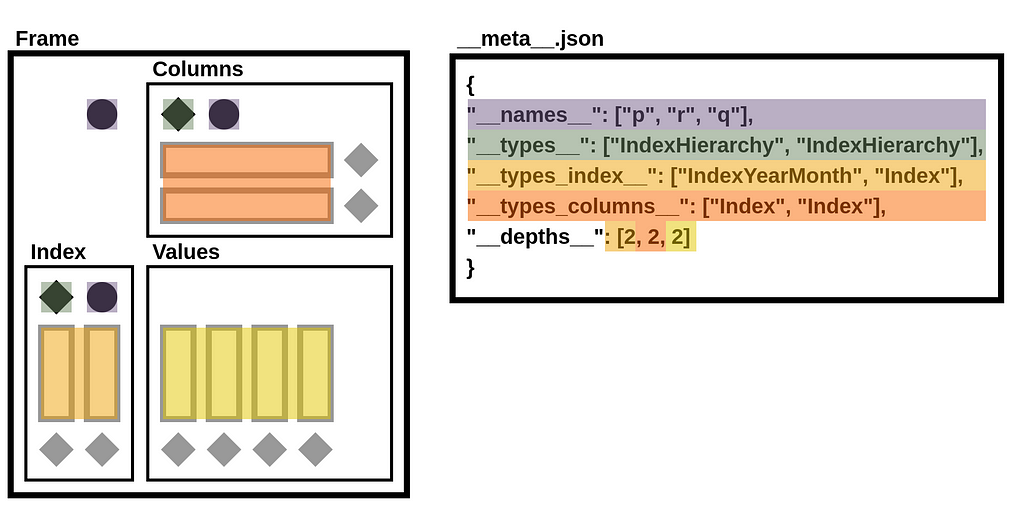
StaticFrame extends the NPZ format to include metadata in a JSON file. This file defines name attributes, component types, and depth counts.
>>> with ZipFile('/tmp/frame.npz') as zf: print(zf.open('__meta__.json').read())
b'{"__names__": ["p", "r", "q"], "__types__": ["IndexHierarchy", "IndexHierarchy"], "__types_index__": ["IndexYearMonth", "Index"], "__types_columns__": ["Index", "Index"], "__depths__": [2, 2, 2]}'
In the illustration below, components of the __meta__.json file are mapped to components of the DataFrame diagram.
As a simple ZIP file, tools to extract the contents of a StaticFrame NPZ are ubiquitous. On the other hand, the ZIP format, given its history and broad features, incurs performance overhead. StaticFrame implements a custom ZIP reader optimized for NPZ usage, which contributes to the excellent read performance of NPZ.
Conclusion
The performance of DataFrame serialization is critical to many applications. While Parquet has widespread support, its generality compromises type specificity and performance. StaticFrame NPZ can read and write DataFrames up to ten-times faster than Parquet with or without compression, with similar (or only modestly larger) file sizes. While Feather is an attractive alternative, NPZ read performance is still generally twice as fast as Feather. If data I/O is a bottleneck (and it often is), StaticFrame NPZ offers a solution.
Faster DataFrame Serialization was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
Originally appeared here:
Faster DataFrame Serialization
