Rodrigo M Carrillo Larco, MD, PhD
A practical solution with code and workflow
Lost in a maze of datasets and endless data dictionaries? Say goodbye to tedious variable hunting! Discover how to quickly identify and extract the variables you need from multiple SAS files using two simple R functions. Streamline your workflow, save time, and make data preparation a breeze!

As a researcher with over seven years of experience working with health data, I’ve often been handed folders full of datasets. For example, imagine opening a folder containing 56 SAS files, each with unique data (example below). If you’ve been in this situation, you know the frustration: trying to locate a specific variable in a sea of files feels like looking for a needle in a haystack.
At first glance, this may not seem like an issue if you already know where your variables of interest are. But often, you don’t. While a data dictionary is usually provided, it’s frequently a PDF document that lists variables across multiple pages. Finding what you need might involve searching (Ctrl+F) for a variable on page 100, only to realize the dataset’s name is listed on page 10. Scrolling back and forth wastes time.
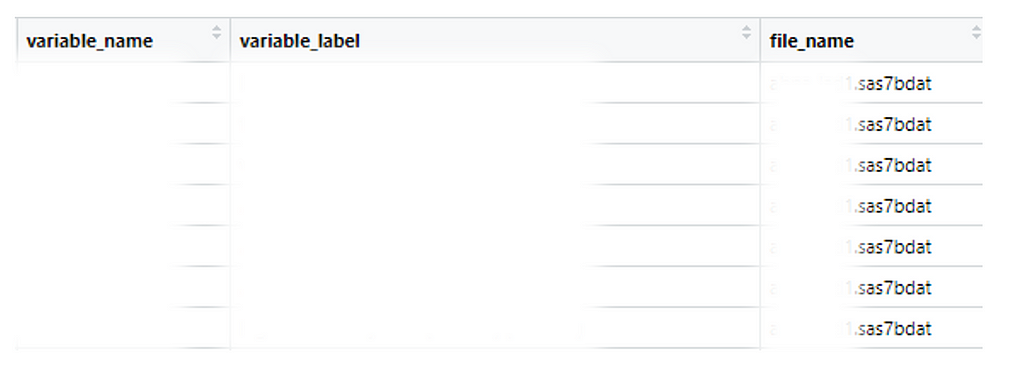
To save myself from this tedious process, I created a reproducible R workflow to read all datasets in a folder, generate a consolidated dataframe of variable names and their labels (example below), and identify where each variable is located. This approach has made my work faster and more efficient. Here’s how you can do it, step by step.
Step-by-Step Guide
Step 1: Use the get_names_labels Function
First, use the custom function get_names_labels (code provided at the end of this post). This function requires the folder path where all your datasets are stored.
path_file <- "D:/folder1/folder2/folder3/folder_with_datasets/"
get_names_labels(path_file)
Step 2: Generate a Variable Dictionary
The get_names_labels function will create a dataframe named names_labels (like the example above), which includes:
· Variable name (variable_name)
· Variable label (variable_label)
· The name of the dataset(s) where the variable was found (file_name)
Depending on the number and size of the datasets, this process may take a minute or two.
Step 3: Search for Variables
Once the names_labels dataframe is generated, you can search for the variables you need. Filter the variable_name or variable_label columns to locate relevant terms. For example, if you’re looking for gender-related variables, they might be labeled as sex, gender, is_male, or is_female.
Be mindful that similar variables might exist in multiple datasets. For instance, age could appear in the main questionnaire, a clinical dataset, and a laboratory dataset. These variables might look identical but differ based on how and where the data was collected. For example:
· Age in the main questionnaire: Collected from all survey participants.
· Age in clinical/lab datasets: Limited to a subset invited for further assessments or those who agreed to participate.
In such cases, the variable from the main questionnaire might be more representative of the full population.
Step 4: Identify Relevant Datasets
Once you’ve determined which variables you need, filter the names_labels dataframe to identify the original datasets (file_name) containing them. If a variable appears in multiple datasets (e.g., ID), you’ll need to identify which dataset includes all the variables you’re interested in.
# Say you want these two variables
variables_needed <- c('ID', 'VAR1_A')
names_labels <- names_labels[which(names_labels$variable_name %in% variables_needed), ]
If one of the variables can be found in multiple original datasets (e.g., ID), you will filter names_labels to keep only the original dataset with both variables (e.g., ID and VAR1_A). In our case, the names_labels dataframe will be reduced to only two rows, one for each of the two variables we were looking for, both of which will be found in the same original dataset.
names_labels <- names_labels %>%
group_by(file_name) %>%
mutate(count = n()) %>%
filter(count >= 2)
Step 5: Extract the Data
Now, use the read_and_select function (provided at the end). Pass the name of the original dataset containing the variables of interest. This function creates a new dataframe in your R environment with only the selected variables. For example, if your variables are in ABC.sas7bdat, the function will create a new dataframe called ABC with just those variables.
unique(names_labels$file_name) # Sanity check, that there is only one dataframe
read_and_select(unique(names_labels$file_name)[1])
Step 6: Clean Your Environment
To keep your workspace tidy, remove unnecessary elements and retain only the new dataframe(s) you need. For example, if your variables of interest came from ABC.sas7bdat, you’ll keep the filtered dataframe ABC which was the output of the read_and_select function.
length(unique(names_labels$file_name))
names_labels$file_name <- str_extract(names_labels$file_name, "[^.]+")
rm(list = setdiff(ls(), c(unique(names_labels$file_name))))
Step 7: Merge Multiple Datasets (Optional)
If your variables of interest are in more than one dataset (e.g., ABC and DEF), you can merge them. Use a unique identifier, such as ID, to combine the datasets into a single dataframe. The result will be a unified dataframe with all the variables you need. You will get a df dataframe with all the observations and only the variables you needed.
# Get a list with the names of the dataframes in the environment (“ABC” and “DEF”)
object_names <- ls()
# Get a list with the actual dataframe
object_list <- mget(object_names)
# Reduce the dataframes in the list (“ABC” and “DEF”) by merging conditional on the unique identifier (“ID”)
df <- Reduce(function(x, y) merge(x, y, by = "ID", all = TRUE), object_list)
# Clean your environment to keep only the dataframes (“ABC” and “DEF”) and a new dataframe “df” which will contain all the variables you needed.
rm(object_list, object_names)
Why This Workflow Works?
This approach saves time and organizes your work into a single, reproducible script. If you later decide to add more variables, simply revisit steps 2 and 3, update your list, and rerun the script. This flexibility is invaluable when dealing with large datasets. While you’ll still need to consult documentation to understand variable definitions and data collection methods, this workflow reduces the effort required to locate and prepare your data. Handling multiple datasets doesn’t have to be overwhelming. By leveraging my custom functions like get_names_labels and read_and_select, you can streamline your workflow for data preparation.
Have you faced similar challenges when working with multiple datasets? Share your thoughts or tips in the comments, or give this article a thumbs up if you found it helpful. Let’s keep the conversation going and learn from each other!
Below are the two custom functions. Save them in an R script file, and load the script into your working environment whenever needed. For example, you could save the file as _Functions.R for easy access.
# You can load the functions as
source('D:/Folder1/Folder2/Folder3/_Functions.R')
library(haven)
library(tidyverse)
library(stringr)
## STEPS TO USE THESE FUNCTIONS:
## 1. DEFINE THE OBJECT 'PATH_FILE', WHICH IS A PATH TO THE DIRECTORY WHERE
## ALL THE DATASETS ARE STORED.
## 2. APPLY THE FUNCTION 'get_names_labels' WITH THE PATH. THE FUNCTION WILL
## RETURN A DATAFRAME NAMES 'names_labels'.
## 3. THE FUNCTION WILL RETURN A DATASET ('names_labels) SHOWING THE NAMES OF
## THE VARIABLES, THE LABELS, AND THE DATASET. VISUALLY/MANUALLY EXPLORE THE
## DATASET TO SELECT THE VARIABLES WE NEED. CREATE A VECTOR WITH THE NAMES
## OF THE VARIABLES WE NEED, AND NAME THIS VECTOR 'variables_needed'.
## 4. FROM THE DATASET 'names_labels', KEEP ONLY THE ROWS WITH THE VARIABLES WE
## WILL USE (STORED IN THE VECTOR 'variables_needed').
## 5. APPLY THE FUNCTION 'read_and_select' TO EACH OF THE DATASETS WITH RELEVANT
## VARIABLES. THIS FUNCTION WILL ONLY NEED THE NAME OF THE DATASET, WHICH IS
## STORED IN THE LAST COLUMN OF DATASET 'names_labels'.
### FUNCTION TO 1) READ ALL DATASETS IN A FOLDER; 2) EXTRACT NAMES AND LABELS;
### 3) PUT NAMES AND LABELS IN A DATASET; AND 4) RETURN THE DATASET. THE ONLY
### INPUT NEEDED IS A PATH TO A DIRECTORY WHERE ALL THE DATASETS ARE STORED.
get_names_labels <- function(path_file){
results_df <- list()
sas_files <- c(
list.files(path = path_file, pattern = "\.sas7bdat$")
)
for (i in 1:length(sas_files)) {
print(sas_files[i])
# Read the SAS file
sas_data <- read_sas(paste0(path_file, sas_files[i]))
sas_data <- as.data.frame(sas_data)
# Get the variable names and labels
var_names <- names(sas_data)
labels <- sas_data %>%
map(~attributes(.)$label) %>%
map_chr(~ifelse(is.null(.), NA, .))
# Combine the variable names and labels into a data frame
var_df <- data.frame(
variable_name = var_names,
variable_label = labels,
file_name = sas_files[i],
stringsAsFactors = FALSE
)
# Append the results to the overall data frame
results_df[[i]] <- var_df
}
results_df <- do.call(rbind, results_df)
#return(results_df)
assign('names_labels', results_df, envir = .GlobalEnv)
}
################################################################################
### FUNCTION TO READ EACH DATASET AND KEEP ONLY THE VARIABLES WE SELECTED; THE
### FUNCTION WILL SAVE EACH DATASET IN THE ENVIRONMENT. THE ONLY INPUNT IS THE
### NAME OF THE DATASET.
read_and_select <- function(df_file){
df_tmp <- read_sas(paste0(path_file, df_file))
df_tmp <- df_tmp %>%
select(unique(names_labels[which(names_labels$file_name == df_file), ]$variable_name)) %>%
as.data.frame()
assign(str_extract(df_file, "[^.]+"), df_tmp,envir = .GlobalEnv)
}
################################################################################
You can find me on LinkedIn and happy to connect and discuss.
Effortless Data Handling: Find Variables Across Multiple Data Files with R was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
Originally appeared here:
Effortless Data Handling: Find Variables Across Multiple Data Files with R