Discover how LDA helps identify critical data features

This article aims to explore Linear Discriminant Analysis (LDA), focusing on its core ideas, its mathematical implementation in code, and a practical example from manufacturing.
I hope you’re on board. Let’s get started!
Who works with industrial data in practice will be familiar with this situation: The datasets usually have many features, and it is often unclear which of the features are important and which are less. “Important” is a relative term in this context. Often, the goal is to differentiate the datasets from each other, i.e., to classify them. A very typical task is to distinguish good parts from bad parts and to identify the causes (aka features) that lead to the failure of the parts.
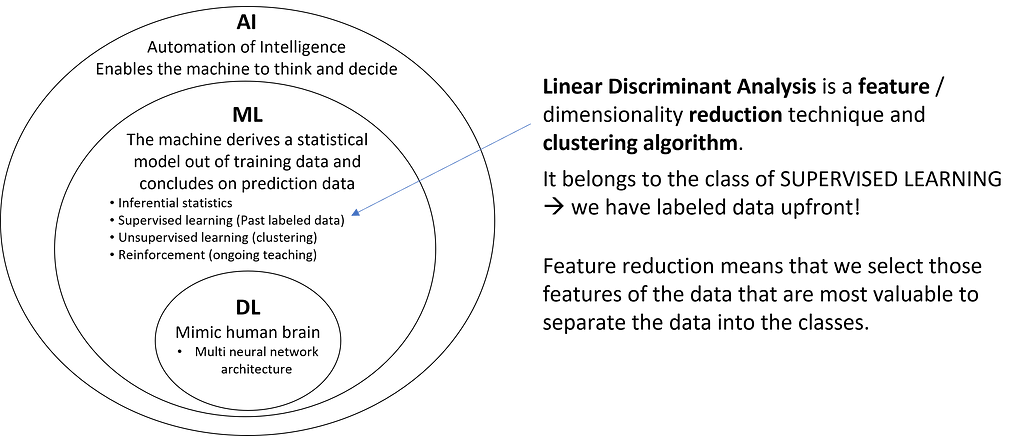
A commonly used method is the well-known Principal Component Analysis (PCA). While PCA belongs to the unsupervised methods, the less widespread LDA is a supervised method and thus learns from labeled data. Therefore, it is particularly suited for explaining failure patterns from large datasets.
1. Goal and Principle of LDA
The goal of LDA is to linearly combine the features of the data so that the labels of the datasets are best separated from each other, and the number of new features is reduced to a predefined count. In AI jargon, this is typically referred to as a projection to a lower-dimensional space.

Excursus: What is dimensionality and what is dimensionality reduction?

Dimensionality refers to the number of features in a dataset.
With just one measurement (or feature), such as the tool temperature from an injection molding machine, we can represent it on a number line. Two features, like temperature and tool pressure, are still manageable: we can easily plot the data on an x-y chart. With three features — temperature, tool pressure, and injection pressure — things get more interesting, but we can still plot the data in a 3D x-y-z chart. However, when we add more features, such as viscosity, electrical conductivity, and others, the complexity increases.

In practice, datasets often contain hundreds or even thousands of features. This presents a challenge because many machine learning algorithms perform poorly as datasets grow too large. Additionally, the amount of data required increases exponentially with the number of dimensions to achieve statistical significance. This phenomenon is known as the “curse of dimensionality.” These factors make it essential to determine which features are relevant and to eliminate the less meaningful ones early in the data science process.
2. How does LDA work?
The process of Linear Discriminant Analysis (LDA) can be broken down into five key steps.
Step 1: Compute the d-dimensional mean vectors for each of the k classes separately from the dataset.
Remember that LDA is a supervised machine learning technique, meaning we can utilize the known labels. In the first step, we calculate the mean vectors mean_c for all samples belonging to a specific class c. To do this, we filter the feature matrix by class label and compute the mean for each of the d features. As a result, we obtain k mean vectors (one for each of the k classes), each with a length of d (corresponding to the d features).

Step 2: Compute the scatter matrices (between-class scatter matrix and within-class scatter matrix).
The within-class scatter matrix measures the variation among samples within the same class. To find a subspace with optimal separability, we aim to minimize the values in this matrix. In contrast, the between-class scatter matrix measures the variation between different classes. For optimal separability, we aim to maximize the values in this matrix.
Intuitively, within-class scatter looks at how compact each class is, whereas between-class scatter examines how far apart different classes are.

Let’s start with the within-class scatter matrix S_W. It is calculated as the sum of the scatter matrices S_c for each individual class:
The between-class scatter matrix S_B is derived from the differences between the class means mean_c and the overall mean of the entire dataset:
where mean refers to the mean vector calculated over all samples, regardless of their class labels.
Step 3: Calculate the eigenvectors and eigenvalues for the ratio of S_W and S_B.
As mentioned, for optimal class separability, we aim to maximize S_B and minimize S_W. We can achieve both by maximizing the ratio S_B/S_W. In linear algebra terms, this ratio corresponds to the scatter matrix S_W⁻¹ S_B, which is maximized in the subspace spanned by the eigenvectors with the highest eigenvalues. The eigenvectors define the directions of this subspace, while the eigenvalues represent the magnitude of the distortion. We will select the m eigenvectors associated with the highest eigenvalues.

Step 4: Sort the eigenvectors in descending order of their corresponding eigenvalues, and select the m eigenvectors with the largest eigenvalues to form a d × m-dimensional transformation matrix W.
Remember, our goal is not only to project the data into a subspace that enhances class separability but also to reduce dimensionality. The eigenvectors will define the axes of our new feature subspace. To decide which eigenvectors to discard for the lower-dimensional subspace, we need to examine their corresponding eigenvalues. In simple terms, the eigenvectors with the smallest eigenvalues contribute the least to class separation, and these are the ones we want to drop. The typical approach is to rank the eigenvalues in descending order and select the top m eigenvectors. m is a freely chosen parameter. The larger m, the less information is lost during the transformation.
After sorting the eigenpairs by decreasing eigenvalues and selecting the top m pairs, the next step is to construct the d × m-dimensional transformation matrix W. This is done by stacking the m selected eigenvectors horizontally, resulting in the matrix W:
The first column of W represents the eigenvector corresponding to the highest eigenvalue, the second column represents the eigenvector corresponding to the second highest eigenvalue, and so on.
Step 5: Use W to project the samples onto the new subspace.
In the final step, we use the d × m-dimensional transformation matrix W, which we composed from the top m selected eigenvectors, to project our samples onto the new subspace:
where X is the initial n × d-dimensional feature matrix representing our samples, and Z is the newly transformed n × m-dimensional feature matrix in the new subspace. This means that the selected eigenvectors serve as the “recipes” for transforming the original features into the new features (the Linear Discriminants): The eigenvector with the highest eigenvalue provides the transformation recipe for LD1, the eigenvector with the second highest eigenvalue corresponds to LD2, and so on.

3. Implementing Linear Discriminant Analysis (LDA) from Scratch
To demonstrate the theory and mathematics in action, we will program our own LDA from scratch using only numpy.
import numpy as np
class LDA_fs:
"""
Performs a Linear Discriminant Analysis (LDA)
Methods
=======
fit_transform():
Fits the model to the data X and Y, derives the transformation matrix W
and projects the feature matrix X onto the m LDA axes
"""
def __init__(self, m):
"""
Parameters
==========
m : int
Number of LDA axes onto which the data will be projected
Returns
=======
None
"""
self.m = m
def fit_transform(self, X, Y):
"""
Parameters
==========
X : array(n_samples, n_features)
Feature matrix of the dataset
Y = array(n_samples)
Label vector of the dataset
Returns
=======
X_transform : New feature matrix projected onto the m LDA axes
"""
# Get number of features (columns)
self.n_features = X.shape[1]
# Get unique class labels
class_labels = np.unique(Y)
# Get the overall mean vector (independent of the class labels)
mean_overall = np.mean(X, axis=0) # Mean of each feature
# Initialize both scatter matrices with zeros
SW = np.zeros((self.n_features, self.n_features)) # Within scatter matrix
SB = np.zeros((self.n_features, self.n_features)) # Between scatter matrix
# Iterate over all classes and select the corresponding data
for c in class_labels:
# Filter X for class c
X_c = X[Y == c]
# Calculate the mean vector for class c
mean_c = np.mean(X_c, axis=0)
# Calculate within-class scatter for class c
SW += (X_c - mean_c).T.dot((X_c - mean_c))
# Number of samples in class c
n_c = X_c.shape[0]
# Difference between the overall mean and the mean of class c --> between-class scatter
mean_diff = (mean_c - mean_overall).reshape(self.n_features, 1)
SB += n_c * (mean_diff).dot(mean_diff.T)
# Determine SW^-1 * SB
A = np.linalg.inv(SW).dot(SB)
# Get the eigenvalues and eigenvectors of (SW^-1 * SB)
eigenvalues, eigenvectors = np.linalg.eig(A)
# Keep only the real parts of eigenvalues and eigenvectors
eigenvalues = np.real(eigenvalues)
eigenvectors = np.real(eigenvectors.T)
# Sort the eigenvalues descending (high to low)
idxs = np.argsort(np.abs(eigenvalues))[::-1]
self.eigenvalues = np.abs(eigenvalues[idxs])
self.eigenvectors = eigenvectors[idxs]
# Store the first m eigenvectors as transformation matrix W
self.W = self.eigenvectors[0:self.m]
# Transform the feature matrix X onto LD axes
return np.dot(X, self.W.T)
4. Applying LDA to an Industrial Dataset
To see LDA in action, we will apply it to a typical task in the production environment. We have data from a simple manufacturing line with only 7 stations. Each of these stations sends a data point (yes, I know, only one data point is highly unrealistic). Unfortunately, our line produces a significant number of defective parts, and we want to find out which stations are responsible for this.
First, we load the data and take an initial look.
import pandas as pd
# URL to Github repository
url = "https://raw.githubusercontent.com/IngoNowitzky/LDA_Medium/main/production_line_data.csv"
# Read csv to DataFrame
data = pd.read_csv(url)
# Print first 5 lines
data.head()

Next, we study the distribution of the data using the .describe() method from Pandas.
# Show average, min and max of numerical values
data.describe()

We see that we have 20,000 data points, and the measurements range from -5 to +150. Hence, we note for later that we need to normalize the dataset: the different magnitudes of the numerical values would otherwise negatively affect the LDA.
How many good parts and how many bad parts do we have?
# Count the number of good and bad parts
label_counts = data['Label'].value_counts()
# Display the results
print("Number of Good and Bad Parts:")
print(label_counts)

We have 19,031 good parts and 969 defective parts. The fact that the dataset is so imbalanced is an issue for further analysis. Therefore, we select all defective parts and an equal number of randomly chosen good parts for the further processing.
# Select all bad parts
bad_parts = data[data['Label'] == 'Bad']
# Randomly select an equal number of good parts
good_parts = data[data['Label'] == 'Good'].sample(n=len(bad_parts), random_state=42)
# Combine both subsets to create a balanced dataset
balanced_data = pd.concat([bad_parts, good_parts])
# Shuffle the combined dataset
balanced_data = balanced_data.sample(frac=1, random_state=42).reset_index(drop=True)
# Display the number of good and bad parts in the balanced dataset
print("Number of Good and Bad Parts in the balanced dataset:")
print(balanced_data['Label'].value_counts())

Now, let’s apply our LDA from scratch to the balanced dataset. We use the StandardScaler from sklearn to normalize the measurements for each feature to have a mean of 0 and a standard deviation of 1. We choose only one linear discriminant axis (m=1) onto which we project the data. This helps us clearly see which features are most relevant in distinguishing good from bad parts, and we visualize the projected data in a histogram.
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler
# Separate features and labels
X = balanced_data.drop(columns=['Label'])
y = balanced_data['Label']
# Normalize the features
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
# Perform LDA
lda = LDA_fs(m=1) # Instanciate LDA object with 1 axis
X_lda = lda.fit_transform(X_scaled, y) # Fit the model and project the data
# Plot the LDA projection
plt.figure(figsize=(10, 6))
plt.hist(X_lda[y == 'Good'], bins=20, alpha=0.7, label='Good', color='green')
plt.hist(X_lda[y == 'Bad'], bins=20, alpha=0.7, label='Bad', color='red')
plt.title("LDA Projection of Good and Bad Parts")
plt.xlabel("LDA Component")
plt.ylabel("Frequency")
plt.legend()
plt.show()
# Examine feature contributions to the LDA component
feature_importance = pd.DataFrame({'Feature': X.columns, 'LDA Coefficient': lda.W[0]})
feature_importance = feature_importance.sort_values(by='LDA Coefficient', ascending=False)
# Display feature importance
print("Feature Contributions to LDA Component:")
print(feature_importance)


The histogram shows that we can separate the good parts from the defective parts very well, with only a small overlap. This is already a positive result and indicates that our LDA was successful.
The “LDA Coefficients” from the table “Feature Contributions to LDA Components” represent the eigenvector from the first (and only, since m=1) column of our transformation matrix W. They indicate the direction and magnitude with which the normalized measurements from the stations are projected onto the linear discriminant axis. The values in the table are sorted in descending order. We need to read the table from both the top and the bottom simultaneously because the absolute value of the coefficient indicates the significance of each station in separating the classes and, consequently, its contribution to the production of defective parts. The sign indicates whether a lower or higher measurement increases the likelihood of defective parts. Let’s take a closer look at our example:
The largest absolute value is from Station 4, with a coefficient of -0.672. This means that Station 4 has the strongest influence on part failure. Due to the negative sign, higher positive measurements are projected towards a negative linear discriminant (LD). The histogram shows that a negative LD is associated with good (green) parts. Conversely, low and negative measurements at this station increase the likelihood of part failure.
The second highest absolute value is from Station 2, with a coefficient of 0.557. Therefore, this station is the second most significant contributor to part failures. The positive sign indicates that high positive measurements are projected towards the positive LD. From the histogram, we know that a high positive LD value is associated with a high likelihood of failure. In other words, high measurements at Station 2 lead to part failures.
The third highest coefficient comes from Station 7, with a value of -0.486. This makes Station 7 the third largest contributor to part failures. The negative sign again indicates that high positive values at this station lead to a negative LD (which corresponds to good parts). Conversely, low and negative values at this station lead to part failures.
All other LDA coefficients are an order of magnitude smaller than the three mentioned, the associated stations therefore have no influence on part failure.
Are the results of our LDA analysis correct? As you may have already guessed, the production dataset is synthetically generated. I labeled all parts as defective where the measurement at Station 2 was greater than 0.5, the value at Station 4 was less than -2.5, and the value at Station 7 was less than 3. It turns out that the LDA hit the mark perfectly!
# Determine if a sample is a good or bad part based on the conditions
data['Label'] = np.where(
(data['Station_2'] > 0.5) & (data['Station_4'] < -2.5) & (data['Station_7'] < 3),
'Bad',
'Good'
)
5. Conclusion
Linear Discriminant Analysis (LDA) not only reduces the complexity of datasets but also highlights the key features that drive class separation, making it highly effective for identifying failure causes in production systems. It is a straightforward yet powerful method with practical applications and is readily available in libraries like scikit-learn.
To achieve optimal results, it is crucial to balance the dataset (ensure a similar number of samples in each class) and normalize it (mean of 0 and standard deviation of 1).
The next time you work with a large dataset containing class labels and numerous features, why not give LDA a try?
Linear Discriminant Analysis (LDA) was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
Originally appeared here:
Linear Discriminant Analysis (LDA)
Go Here to Read this Fast! Linear Discriminant Analysis (LDA)