Customizable R package for Graph / Network visualization
Clinical Flowchart
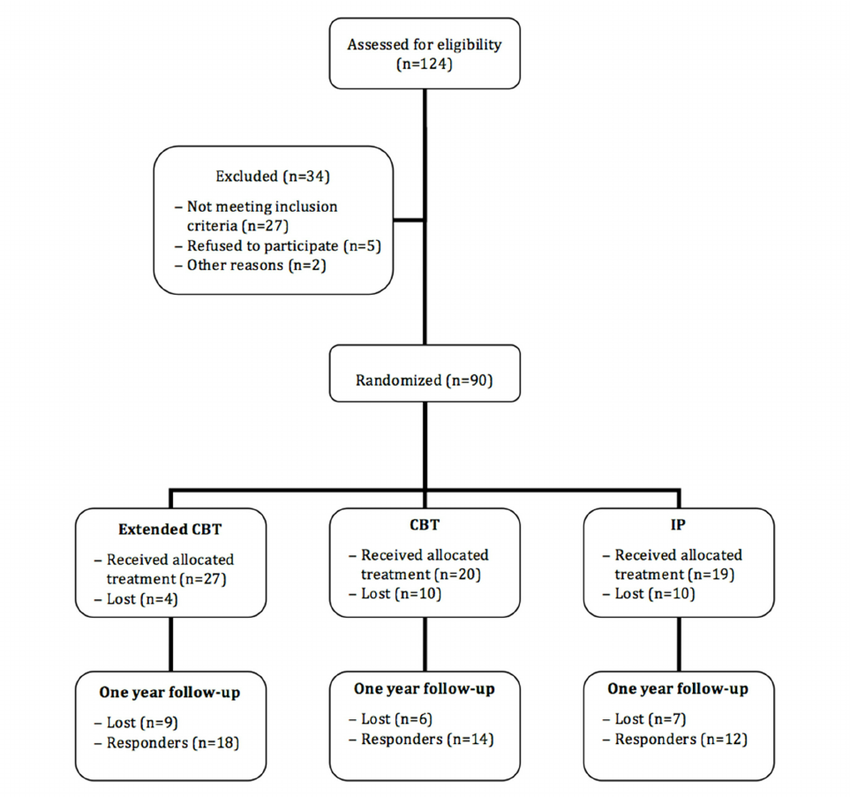
A Clinical (Trial) Flowchart is a visual representation of each step and procedure in a clinical trial or treatment process.
It starts with patients, lists which therapeutic methods are used, which patients are excluded from the trial and for what reason, how the groups are assigned, and so on, and looks like the example below.

This illustration implies the following
- Initially started with 124 patients
- Exclude 34 patients due to ineligibility or declining to participate
- The remaining 90 patients are randomly assigned to a group (Extended CBT, CBT, IP) to compare treatments.
- In each group, 4, 10, and 10 people drop out of the course (although ideally 30 people would be split), and the rest receive the assigned treatment.
- When the patients were followed up after 1 year, 9, 6, and 7 were unresponsive, respectively.
The trial started with 124 patients and ended up with only 44 patients, which shows how difficult and expensive clinical trials can be.
Anyway, there is no set way to draw this flowchart, and you can use any commercial software such as PowerPoint or Keynote, or web-based diagramming tools such as draw.io or lucidchart.
Clinical Flowchart with R
I don’t know exactly why, but this time I needed to use R to plot the chart.
The advantages of using a programming approach like R include automation and reuse, integration with other functions (e.g., a program that plots from data source to chart), and a level of customization not available in commercial programs.
Anyway, here’s how I’ve tried to accomplish this.
Since the original purpose of the package was to draw participant flow diagrams, this is the closest I could get to what I was trying to achieve. I think it’s the best choice unless you have special circumstances.
A library for graph and network visualization that leverages features from d3 and mermaid.
- ggplot2 + ggforce
R’s best friend ggplot (recommended by GPT) and its companion packages.
R package for network visualization graphs based on vis.js.
However, as it turns out, all four methods failed.
This is because there was a special situation with the figure I was trying to draw.
The following figure shows the actual drawing, with only the numbers and groups replaced with 1000, 1,2,3… A, B.

There were two problematic parts: the Completed curative surgery part in the middle, where the two nodes from the previous step connect to one long node, and the difficulty of positioning the edge.

I considered several ways to do this, and eventually decided to use an old friend, shinyCyJS, which I can customize to my purpose.
shinyCyJSis a R package to leverage the network/graph visualization capabilities of cytoscape.js in R. It was the first thing I wrote when I was looking for genomic network visualization when I graduated (at the time, there was only igraph and RCyjs) and didn’t find what I was looking for, which is how I ended up at my current job.
2 Custom feature with shinyCyJS
- Positioning Edges
In order to implement the positioning of the edges in the two custom features above, I initially tried to use taxi edges.
However, again, this only determines the edge based on the position of the node, and it is not possible to move the edge, so I switched to adding a microscopic node in the middle and traversing the edge to it, as shown below (since it is possible to specify the position of the “micro” node).


- One Big Node
In cytoscape.js, by default, nodes are connected by edges that consider the shortest distance between the center and the center, and if they go through other points along the way, like the taxi mentioned above, the points are not specified and are calculated algorithmically.

The problem with allowing intermediate points to be specified is that when there are multiple edges between nodes, like in the bezier and haystack examples, it is annoying to have to specify intermediate points for all of them.
In the previous example, there are only three edges, so it’s not a big deal, but in the case of genomic networks, which I studied in graduate school, a single gene often interacts with dozens or hundreds of other genes, so it’s a big deal.
In the worst case scenario, the edges overlap and missing a few edges can cause the graph to produce completely different information.

In other words, in the problem of connecting to one long node, the long node is only a graphical (width) perspective for the user,
But from the computer’s point of view, it is irrational behavior to connect an edge to a node that doesn’t even exist, as shown in the image below, so there is no reason to consider this option in the first place.

To solve this problem, I created a micro node and modified it to connect to the appropriate part, just like the previous edge midpoint problem.

Here is a partial view of the graph I ended up creating in R. (Again, groups and numbers are arbitrarily modified)

Another problem, download as PNG
Technically, you can use the Export as PNG button in Rstudio viewer as shown below, and if that doesn’t work, you can take a screenshot, but cytoscape.js has a function to save the graph as png, so I used it.

*I actually had a request to add a download to png feature to shinyCyJS a while back, and I replied “why not just take a screenshot?”.
This required using an internet browser (chrome) (As cytoscape.js is Javascript) and that meant I had to go beyond R and implement it on the web using Shiny.
Of course, shinyCyJS is a package that was built with shiny integration in mind from the start, so it was no problem.
Below is the code you need to run in Chrome’s developer tools to download it
const pngBlob = await cy.png({
output: 'blob-promise',
});
const fileName = 'myfile.png';
const downloadLink = document.createElement('a');
downloadLink.href = URL.createObjectURL(pngBlob);
downloadLink.download = fileName;
downloadLink.click();
shinyCyJS is an R package I wrote, and it literally does everything cytoscape does, plus custom features like this if needed, so if you need to do network/graph visualization in R, you can try it out or ask for what you need.
Of course, if you don’t need to use R, draw.io seems better.
Additionally, if you want to package other Javascript libraries for use with R, you can send me an email.
Thanks for reading.
Use R to build Clinical Flowchart with shinyCyJS was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
Originally appeared here:
Use R to build Clinical Flowchart with shinyCyJS
Go Here to Read this Fast! Use R to build Clinical Flowchart with shinyCyJS