An outlier detection method that determines a relevant distance metric between records
Outliers are often defined as the items in a dataset that are very different than the majority of the other items. That is: any record that is significantly different from all other records (or from almost all other records), and is more different from the other records than is normal, could reasonably be considered an outlier.

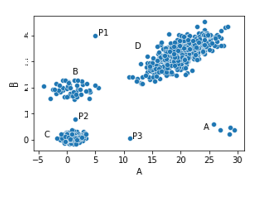
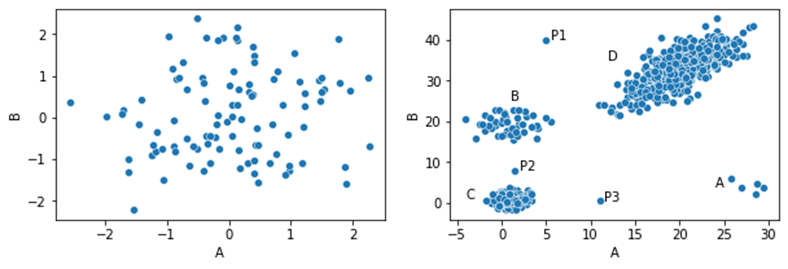
In the dataset shown here, we have four clusters (A, B, C, and D) and three points outside these clusters: P1, P2, and P3. These can likely be considered outliers, as they are each far from all other points — that is, they are significantly different than most other points.
As well, Cluster A has only five points. While these points are fairly close to each other, they are far from all other points, so could quite possibly be considered outliers as well — again, based on the distances from these points to the majority of other points.
The inliers, on the other hand (the points within the larger clusters), are all very close to a significant number of other points. For example, any point in the middle of Cluster C is very close to many other points (i.e. is very similar to many other points), so would not be considered an outlier.
There are numerous other ways we can look at outliers, and many other approaches are actually used for outlier detection — for example outlier detection methods based on Frequent Item Sets, Association Rules, compression, Markov Models, and so on. But identifying the records that are similar to few other records, and that are relatively different from the records they are most similar to, is very common. This is, in fact, the underlying idea behind many of the most common outlier detection algorithms, including kNN, LOF (Local Outlier Factor), Radius, and numerous other algorithms.
But, using this approach leaves the question of how to quantify how different a record is from the other records. There are a number of techniques to do this. Some of the most common in outlier detection include Euclidean, Manhattan, and Gower distances, as well as a number of similar metrics.
We’ll cover these quickly below. But we want to look in this article specifically at a very versatile, and likely under-used, method for calculating the difference between two records in tabular data that’s very useful for outlier detection, called Distance Metric Learning — as well as a method to apply this specifically to outlier detection.
This article continues a series on outlier detection that includes Counts Outlier Detector, Frequent Patterns Outlier Factor, and techniques to tune and test detectors (using a method called doping). It also includes another excerpt from my book Outlier Detection in Python.
Distance Metrics
To determine if a record is 1) unusually far from most other records; and 2) close to relatively few records, we generally first calculate the pairwise distances: the distances between each pair of records in a dataset. In practice, we may take a more optimized approach (for example only calculating approximate distances where records are known to be very far apart in any case), but, at least in principle, calculating the distances between each pair of rows is common in outlier detection.
Which means, we need a way to calculate the distance between any two records.
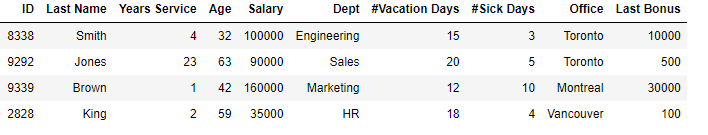
If we have a set of data such as the following, a large table of staff records (here showing a random subset of four rows), how can we best say how similar any two rows are?
Euclidean Distances
One very common method is to use the Euclidean distance.
Before looking further at the staff data, consider again the scatter plot above. We see here a case where using the Euclidean distance feels natural. As this dataset contains only two features, and both are numeric, plotting the data as in this figure (as a scatter plot) is fairly intuitive. And, once plotted in this way, we naturally picture the Euclidean distances between points (based on the Pythagorean formula).
In cases, though, with: many features; where many of these are categorical; and with associations among the columns, the Euclidean distances between rows, while still valid and often useful, can feel less natural
An issue with Euclidean distances is that they are really intended for numeric data, though most real-world data, like the staff records, is mixed: containing both numeric and categorical features. Categorical values can be encoded numerically (using, for example, One-Hot, Ordinal, or other encoding methods), which then allows calculating Euclidean distances (as well as other numeric distance measures). But it isn’t always ideal. And each method of numeric encoding has its own implications for the distances calculated. But it is quite possible, and quite common.
Considering the Staff table above: we would likely leave ID and Last Name out of the outlier detection process, using the remainder of the columns. Given that, we will still have the Department and Office features as categorical. Let’s assume we encode these using one-hot encoding.
To calculate the Euclidean distances between rows, we also must scale the numeric features, putting all features on the same scale. This can be done a variety of ways, include Standardizing (converting values to their z-values, based on the number of standard deviations a value is from the mean of that column), or min-max scaling.
Once the data is numerically encoded and scaled, we may then calculate the Euclidean distance between every pair of rows.
Gower Distances
Alternatively, given we have some categorical features, we can use a method designed for mixed data, such as the Gower distance. This, to compare any two rows, takes the difference column by column and sums these differences. Where the data is strictly numeric, it is equivalent to the Manhattan distance.
For categorical columns, with Gower distances, usually Ordinal Encoding is used, as we are only concerned if there is an exact match or not. The difference in two values of a categorical column is then either 0.0 or 1.0. In the Staff table above, Smith and Jones have a distance of 1.0 for Department (1.0 is always used with different values: ‘Engineering’ and ‘Sales’ in this case) and a distance of 0.0 for Office (0.0 is always used where two rows have the same value: ‘Toronto’ in this case).
To compare the numeric fields, as with Euclidean, and most distance metrics, we will need to scale them first, so that the numeric fields may all be treated equally. As indicated, there are a number of ways to do this, but let’s assume we use min-max scaling here, which puts all values on a scale between 0.0 and 1.0. We may then have a table such as:
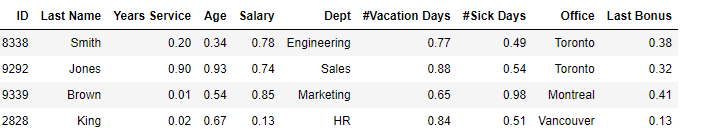
The difference (using Gower Distance) between Smith and Jones would then be: abs(0.90 — 0.20) + abs(0.93 — 0.34) + abs(0.74 — 0.78) + 1.0 + abs(0.88 — 0.77) + abs(0.54 — 0.49) + 0.0 + abs(0.32 — 0.38).
That is, skipping ID and Last Name, we calculate the absolute difference in each numeric field and take either 0.0 or 1.0 for each categorical field.
This may be reasonable, though does have some issues. The main one is likely that the categorical fields have more weight than the numeric: they will often have a difference of 1.0, where numeric values will tend to have smaller differences. For example, the Age difference between Smith and Jones is quite large, but will only have a difference of abs(0.93–0.34), or 0.59 (still significant, but less than the 1.0 that the Department counts towards the total difference between the rows). As covered in Outlier Detection in Python, one-hot encoding and other encodings with other distance metrics have similar issues handling mixed data.
As well, all categorical features are equally relevant as each other; and all numeric features are equally relevant as each other, even where some are, for example, highly correlated, or otherwise should possibly carry more or less weight.
In general, distance metrics such as Euclidean or Gower distance (and other metrics such as Manhattan, Canberra and so on), may be appropriate distance metrics in many cases, and are often excellent choices for outlier detection. But, at the same time, they may not always be ideal for all projects.
Euclidean Distances Viewed as Physical Distances in High Dimensional Space
Looking again at Euclidean distances, these essentially consider the records each as points in high-dimensional space, and calculate the distances between these points in this space. Manhattan and Gower distances are a bit different, but work quite similarly.
As as simpler example than the full Staff table, consider this table, but for the moment just including the numeric features: Years of Service, Age, Salary, # Vacation Days, # Sick Days, and Last Bonus. That’s six features, so each row can be viewed as a point in 6-dimensional space, with the distances between them calculated using the Pythagorean formula.
This is reasonable, but is certainly not the only way to look at the distances. And, the distance metric used can make a substantial difference to the outlier scores assigned. For example, Euclidean distances can put more emphasis on a few features with very different values than, say, Manhattan distances would.
Example of Euclidean and Manhattan Distances
We’ll consider here two different cases of this 6-dimensional data (showing also the ID and Last Name columns for reference).
First, an example for two staff, Greene and Thomas, where most values are similar, but Years Service is very different:
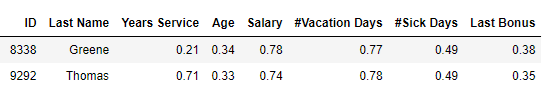
Second, an example for two other staff, Ford and Lee, with most values moderately different but none very different:
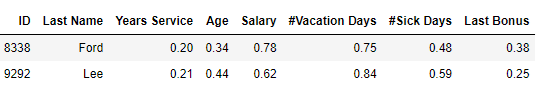
Which of these pairs of rows is most similar? Using Manhattan distances, Greene and Thomas are most similar (having a distance of 0.59, compared to 0.60). Using Euclidean distances, Ford and Lee are most similar (having a distance of 0.27, compared to 0.50).
It’s not always clear when using Manhattan or Euclidean distances is more suitable, or when it’s preferable to use another metric, such as Canberra, or Minkowski (using, for example, cubed distances), Mahalanobis, and so on. This is not necessarily an issue, but it does highlight that there’s many ways to look at the distances between rows.
Euclidean distances, specifically, imply we’re viewing the data as points in high-dimensional space, and are taking what’s equivalent to the physical distance between them. This has some real value, but it isn’t always entirely natural. Simply looking at a table of values, such as the Staff data above, we picture the rows (in this example) as staff records, not points in space.
And, using the Euclidean distance requires taking the squared age, squared salary, and so on — which lacks any intuitive appeal. It’s not clear what something like, for example, the squared age really means. It can work well, but a geometric interpretation of the data is really just one of many ways we can picture the data.
Further, it’s a generic method, that does not consider the data itself.
Distance Metric Learning
Distance Metric Learning presents another way to think about the problem of determining how similar two records are. Instead of first defining a distance measure and then applying it to the data at hand, Distance Metric Learning attempts to learn from the data itself how similar records are to each other.
It also addresses a limitation of Euclidean, Manhattan, and most other distance metrics: that all features are treated equally, whether this is most appropriate or not.
The idea here is: some features are more relevant than others, and some features are related to each other (in some cases, sets of features may even be redundant, or nearly). Simply treating every feature identically is not necessarily the best way to identify the most anomalous records in a dataset.
Distance Metric Learning is a major area in itself, but I’ll cover here one approach to how it may be applied to outlier detection. Specifically, we’ll look here at an application Distance Metric Learning for outlier detection based on creating Random Forests.
Assume, for the moment, that:
- We have a Random Forest that predicts some target
- We have a table of data that can be passed through the Random Forest (e.g. the staff data, but any tabular data)
- We want to calculate the distances between each pair of rows.
We’ll use these pairwise distances for outlier detection for the discussion here, but could in principle use them for any purpose.
We’ll describe soon how to create a Random Forest for this, but assume for the moment that we have a Random Forest and that it is of good quality, well-trained, and robust.
One thing we can do to estimate how similar rows are to each other is look at the predictions the Random Forest makes. Let’s assume the Random Forest is trained as a binary classifier, so can produce, for each record in the data, a predicted probability of being the positive class.
Two records passed through the Random Forest may have very similar probabilities, say 0.615 and 0.619. These are very close, so we can suspect that the two records are similar to each other. But, not necessarily. They may actually follow quite different decision paths through the many decision trees within the Random Forest, and happen to average out to similar predictions. That is, they may receive similar predictions for different reasons, and may not be similar at all.
What’s most relevant is the decision paths the records take through the decision trees. If two records take the same paths in most of the trees (and so end in the same leaf nodes), then we can say that they are similar (at least in this respect). And if they, for the most part, end in different leaf nodes, we can say they are different.
This, then, provides a powerful tool to determine, in a sensible way, how similar any two records are.
Creating a Random Forest
This is clearly a useful idea, but it does require a Random Forest, and a Random Forest that is meaningful for this purpose — one that captures well the nature of the data available.
One way to create such a Random Forest is to build one that learns to distinguish this data from similar, but fake, data. That is, data that’s synthetically generated to be similar, but not quite the same as this data (such that it is distinguishable).
So, if we can create a such a set of fake data, we can then train a Random Forest classifier to distinguish the two types of data.
There are a number of ways to create the synthetic data to be used here, including several covered in Outlier Detection in Python. One, for example, is doping (also covered in this Medium article). We’ll look, though, at another method here that can work well. This can be overly simplistic and not always as effective as more sophisticated techniques, but it does provide a nice, simple introduction to the idea.
Here we generate an equal number of synthetic records as there are real records. An exactly balanced set isn’t necessary and some imbalance may actually work better in some cases, but this example, for simplicity, uses a balanced dataset.
We generate the synthetic data one row at a time, and for each row, one feature at a time. To generate a value, if the feature is categorical, we select a value from the real data with a probability proportional to the distribution in the real data. For example, if the real data contains a column for Colour and this contains 450 rows with Red, 650 rows with Blue, 110 rows with Green, and 385 rows with Yellow, then, as fractions these are: Red: 0.28, Blue: 0.41, Green: 0.07, Yellow: 0.24. A set of new values will be created for this column in the synthetic data with similar proportions.
If the feature is numeric, we calculate the mean and standard deviation of the real data for this feature and select a set of random values from a Normal distribution with these parameters. Any number of other ways to do this may be considered as well, but again, this is a straightforward introduction to the idea.
Doing this we generate synthetic data where each row is comprised entirely of realistic values (each row can potentially contain rare values in categorical columns, and potentially rare or extreme values in numeric columns — but they are all reasonably realistic values).
But, the normal relationships between the features are not respected. That is: as each column value is generated independently, the combination of values generated may be unrealistic. For example if creating synthetic data to mimic the Staff table above, we may create fake records that have an Age of 23 and Years of Service of 38. Both values, on their own, are realistic, but the combination is nonsensical and, as such, should be an unseen combination in the real data — so distinguishable from the real data.
The synthetic data for numeric fields can be created with code (in python) such as:
real_df['Real'] = True
synth_df = pd.DataFrame()
for col_name in real_df.columns:
mean = real_df[col_name].mean()
stddev = real_df[col_name].std()
synth_df[col_name] = np.random.normal(
loc=mean, scale=stddev, size=len(real_df))
synth_df['Real'] = False
train_df = pd.concat([real_df, synth_df])
Here, we assume the dataframe real_df contains the real data. We then create a second dataframe called synth_df, then combine both into train_df, which can be used to train a Random Forest to distinguish the two.
Categorical data can be created similarly:
for col_name in real_df.columns:
vc = real_df[col_name].value_counts(normalize=True)
synth_df[col_name] = np.random.choice(a=vc.keys().tolist(),
size=len(real_df),
replace=True,
p=vc.values.tolist())
As indicted, this is only one way to generate the data, and it may be useful to tune this process, allowing more unusual single values, or restricting to less unusual relationships among the features.
Once this data is created, we can train a Random Forest to learn to distinguish the real from the fake data.
Once this is done, we can actually also perform another form of outlier detection as well. Any real records that are passed through the Random Forest, where it predicts this record is fake, may be considered anomalous — they are more similar to the synthetic data than the real data. This is covered in Outlier Detection in Python, but for this article, we’ll focus on Distance Metric Learning, and so look at the decision paths through the trees within the Random Forest (and not the final predictions).
Using the Random Forest to Measure Outlierness
As described above, if two records tend to end in almost entirely different leaf nodes, they can be considered different, at least in this sense.
It’s possible to, for each pair of records, count the number of trees within the Random Forest where they end in the same leaf node and where they end in different leaf nodes. But, there’s also a simpler method we can use. For each record passed through the Random Forest, for each tree, we can see what the terminal (leaf) node is. We can also see how many records in the training data ended in that node. The fewer training records, the more unusual this path is.
If, over most trees, a record ends in the same leaf nodes as very few other records, it can be considered anomalous.
The main idea is: if the Random Forest is accurate, it can distinguish real from fake records well. So, when passing a real record through the Random Forest, it will likely end in a leaf node associated with the real data. If it is a normal real record, it will follow a common path, used by many other real records. At each step on the path, the node in the Decision Tree will split on one feature — a feature and split point that is effective at separating real from synthetic data. A typical record will have a value associated with common real data, so will follow the path at each split point associated with real data.
If a Random Forest contained only a small number of trees, the size of the leaf node each record ends in could be quite arbitrary. But, Random Forests can be set to have hundreds or thousands of trees. Where records consistently end in leaf nodes that are unusual for their trees, the record can reasonably be considered anomalous.
There can still be some variance to the process, even where a large Random Forest is used. To address this, instead of using a single Distance Metric Learning outlier detector, it’s possible to use several, combined in an ensemble. That’s beyond the scope of this article, but the general idea is to create a variety of synthetic datasets and for each a variety of Random Forests (with different hyperparameters), then average the results together.
Example
To demonstrate the idea, we’ll create a simple Distance Metric Learning detector.
But first, we’ll create a couple test datasets. These are both numeric datasets with two features. As indicated, this is less realistic than data with many features, and with a number of categorical features, but it is useful for demonstration purposes — it is easy to plot and understand.
The first test set is a single cluster of data:
import numpy as np
import pandas as pd
def create_simple_testdata():
np.random.seed(0)
a_data = np.random.normal(size=100)
b_data = np.random.normal(size=100)
df = pd.DataFrame({"A": a_data, "B": b_data})
return df
The second actually creates the dataset shown at the beginning of the article, with four clusters and three points outside of these.
def create_four_clusters_test_data():
np.random.seed(0)
a_data = np.random.normal(loc=25.0, scale=2.0, size=5)
b_data = np.random.normal(loc=4.0, scale=2.0, size=5)
df0 = pd.DataFrame({"A": a_data, "B": b_data})
a_data = np.random.normal(loc=1.0, scale=2.0, size=50)
b_data = np.random.normal(loc=19.0, scale=2.0, size=50)
df1 = pd.DataFrame({"A": a_data, "B": b_data})
a_data = np.random.normal(loc=1.0, scale=1.0, size=200)
b_data = np.random.normal(loc=1.0, scale=1.0, size=200)
df2 = pd.DataFrame({"A": a_data, "B": b_data})
a_data = np.random.normal(loc=20.0, scale=3.0, size=500)
b_data = np.random.normal(loc=13.0, scale=3.0, size=500) + a_data
df3 = pd.DataFrame({"A": a_data, "B": b_data})
outliers = [[5.0, 40],
[1.5, 8.0],
[11.0, 0.5]]
df4 = pd.DataFrame(outliers, columns=['A', 'B'])
df = pd.concat([df0, df1, df2, df3, df4])
df = df.reset_index(drop=True)
return df
The two datasets are shown here:
We next show a simple outlier detector based on Distance Metric Learning. This detector’s fit_predict() method is passed a dataframe (within which we identify any outliers). The fit_predict() method generates a synthetic dataset, trains and Random Forest, passes each record through the Random Forest, determines which node each record ends in, and determines how common these nodes are.
from sklearn.ensemble import RandomForestClassifier
from collections import Counter
from sklearn.preprocessing import RobustScaler
class DMLOutlierDetection:
def __init__(self):
pass
def fit_predict(self, df):
real_df = df.copy()
real_df['Real'] = True
# Generate synthetic data that is similar to the real data
# For simplicity, this covers just the numeric case.
synth_df = pd.DataFrame()
for col_name in df.columns:
mean = df[col_name].mean()
stddev = df[col_name].std()
synth_df[col_name] = np.random.normal(loc=mean,
scale=stddev, size=len(df))
synth_df['Real'] = False
train_df = pd.concat([real_df, synth_df])
clf = RandomForestClassifier(max_depth=5)
clf.fit(train_df.drop(columns=['Real']), train_df['Real'])
# Get the leaf node each record ends in
r = clf.apply(df)
# Initialize the score for all records to 0
scores = [0]*len(df)
# Loop through each tree in the Random Forest
for tree_idx in range(len(r[0])):
# Get the count of each leaf node
c = Counter(r[:, tree_idx])
# Loop through each record and update its score based
# on the frequency of the node it ends in
for record_idx in range(len(df)):
node_idx = r[record_idx, tree_idx]
node_count = c[node_idx]
scores[record_idx] += len(df) - node_count
return scores
df = create_four_clusters_test_data()
df = pd.DataFrame(RobustScaler().fit_transform(df), columns=df.columns)
clf = DMLOutlierDetection()
df['Scores'] = clf.fit_predict(df)
This code example just runs on the data created by create_four_clusters_test_data(), but can be called with the data from create_simple_testdata() as well.
The results can be visualized with code such as:
import matplotlib.pyplot as plt
import seaborn as sns
sns.scatterplot(x=df["A"], y=df['B'], hue=df['Scores'])
plt.show()
The results of both test datasets are shown below, drawing the original data, but setting the hue by their outlier score (placed in the ‘Scores’ column by the code above).
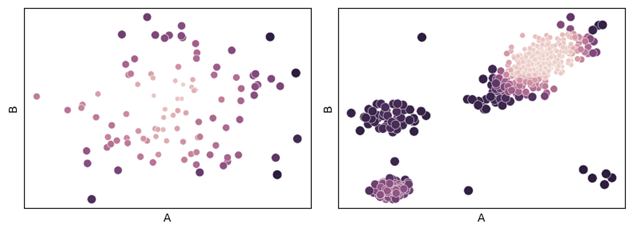
In the dataset on the left, with a single cluster, the outermost points receive the highest scores, which is as expected. In the dataset on the right, with four clusters, the highest outlier scores go to the three points outside the clusters, the smaller clusters, and the points on the edge of the largest clusters. This is quite reasonable, though other detectors may score these differently, and likely also quite reasonably.
As indicated above, using Euclidean distances can be natural for these datasets, though possibly less so for datasets with many features, categorical features, associations between features, and other nuances to the data. But, even in these simpler cases where Euclidean works quite well, Distance Metric Learning can also work well, and provides a natural outlier detection method. Working with more complex data, this can be the case even more so.
Conclusions
Distance Metric Learning can be used for many purposes outside of outlier detection, and even within outlier detection, can be used a variety of ways. For example, it’s possible to use a Random Forest as above to calculate the pairwise distances in a dataset and pass these to another algorithm. DBSCAN, for example, provides a ‘precomputed’ option, which allows passing a pre-calculated matrix of pairwise distances; it’s then possible to use DBSCAN (or a similar clustering method, such as HDBSCAN) for one of several possible clustering-based outlier detection algorithms.
And, Distance Metric Learning can also be used, as in this article, in a more direct way, which is an excellent outlier detection method in itself. In many cases, it can be favorable for detecting outliers than methods based on Euclidean, Manhattan, Gower, or other such distance metrics. It can also provide diversity to an ensemble of detectors, even where these methods also work well.
No outlier detection method is definitive, and it’s generally necessary to use multiple outlier detection methods on any given project (including, often, the same method multiple times, using different parameters), combining their results to achieve strong overall outlier detection.
So, Distance Metric Learning won’t work for every project and where it does it may (as with any detector) perform best when combined with other detectors. But, this is a valuable tool; Distance Metric Learning can be a very effective technique for outlier detection, though it receives less attention than other methods.
It does require some tuning, both in terms of how the synthetic data is produced and in terms of the hyper-parameters used by the Random Forest, but once tuned, provides a strong and intuitive outlier detection method.
All images by the author
Distance Metric Learning for Outlier Detection was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
Originally appeared here:
Distance Metric Learning for Outlier Detection
Go Here to Read this Fast! Distance Metric Learning for Outlier Detection