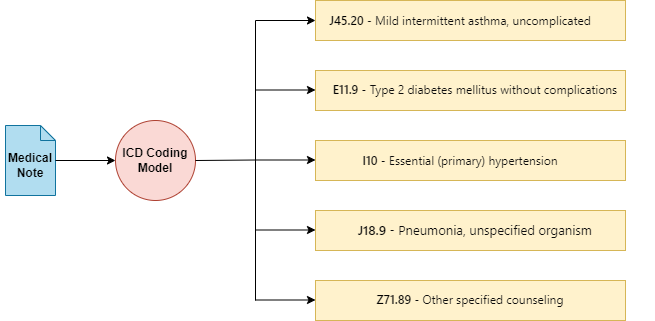
Exploring LLMs for ICD Coding — Part 1
Building automated clinical coding systems with LLMs
Clinical coding isn’t common parlance, but it significantly impacts everyone who interacts with the healthcare system in most countries. Clinical coding involves translating and mapping medical information from patient health records, such as diagnoses and procedures, into standardized numeric or alphanumeric codes. These codes are crucial for billing, healthcare analytics, and ensuring that patients receive appropriate care.

Clinical coding is typically performed by human coders with medical expertise. These coders navigate complex and often hierarchical coding terminologies with specific codes for a vast range of diagnoses and procedures. As such, coders must have a deep familiarity with and experience in the coding terminology used. However, manually coding documents can be slow, error-prone, and bottlenecked by the requirement for significant human expertise.
Deep learning can play a significant role in automating clinical coding. By automating the extraction and translation of complex medical information into codes, deep learning systems can function as a valuable tool within a human-in-the-loop system. They can support coders by processing large volumes of data quickly, potentially improving speed and accuracy. This can help streamline administrative operations, reduce billing errors and enhance patient care outcomes.
In this first part, I describe what ICD coding is, characterize the various challenges that an automated coding system must overcome in order to be effective. I also analyze how Large Language Models (LLMs) can be effectively used for overcoming these problems, and illustrate that by implementing an algorithm from a recent paper that leveraged LLMs effectively for ICD coding.
Table of Contents:
- What is ICD Coding?
- What are the challenges in automated ICD coding?
- How can LLMs help in automated ICD coding?
- Exploring the paper “Automated clinical coding using off-the-shelf large language models
- Implementing the technique described in the paper
- Conclusion
- References
What is ICD Coding?
The International Classification of Diseases (ICD) coding is a clinical terminology system developed and maintained by the World Health Organization [1]. It is used in most countries to categorize and code all diagnoses, symptoms, and procedures recorded for a patient.
Medical notes, which document the diagnoses and medical procedures for a patient, are crucial for ICD coding . The ICD terminology features a hierarchical, tree-like structure to efficiently organize extensive information, with approximately 75,000 different codes available for various medical conditions and diagnoses. Coding these documents precisely is vital; accurate coding ensures appropriate billing and influences the quality of healthcare analysis, directly impacting patient care outcomes, reimbursement and healthcare efficiency.
What are the challenges in automated ICD coding?
ICD coding poses multiple challenges that an automated system must overcome in order to be effective.
Label Diversity in ICD Coding:
One significant challenge is the extensive output space of labels. ICD codes are numerous, and each code can differ in minute details — for instance, a condition affecting the right hand versus the left hand will have different codes. Additionally, there exists a long tail of rare codes that appear infrequently in medical records, making it difficult for deep learning models to learn and accurately predict these codes due to the scarcity of examples.
Adapting to New ICD Codes:
Traditional datasets used for training, such as MIMIC-III [2] , while comprehensive, often limit the scope of ICD codes to those included in the training corpus. This restriction means that deep-learning models treating ICD coding as a multi-label classification problem from medical notes to ICD codes have difficulty handling new codes introduced into the ICD system after the model’s training. This makes retraining necessary and potentially challenging.
Extracting and Contextualizing Information:
Another major challenge is the accurate extraction and contextualization of information from medical notes. ICD coding is fundamentally an Information Retrieval problem that requires not only identifying the diagnoses in the medical records but also capturing all supplementary information necessary for correctly mapping these diagnoses to their respective ICD codes. Therefore, it is crucial for an automated system to extract the various medical diagnoses in the medical note and contextualize them appropriately to ensure accurate mapping to the ICD codes.
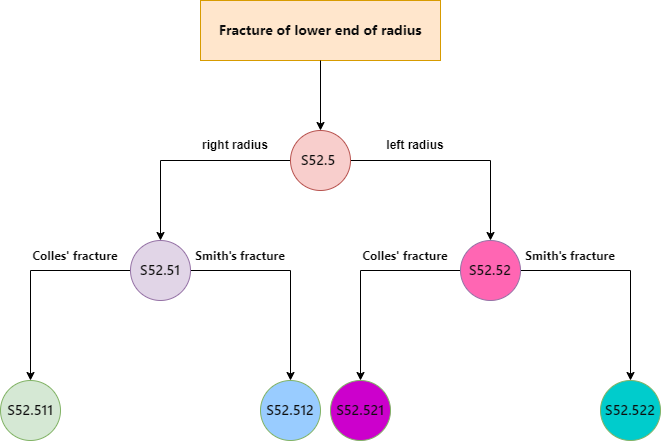
What does contextualization mean here? When dealing with medical notes, to contextualize a diagnosis means to link it with all pertinent details — such as the bodypart affected and the symptoms of the condition — to fully characterize the diagnosis. Generally this task is referred to as relation extraction.
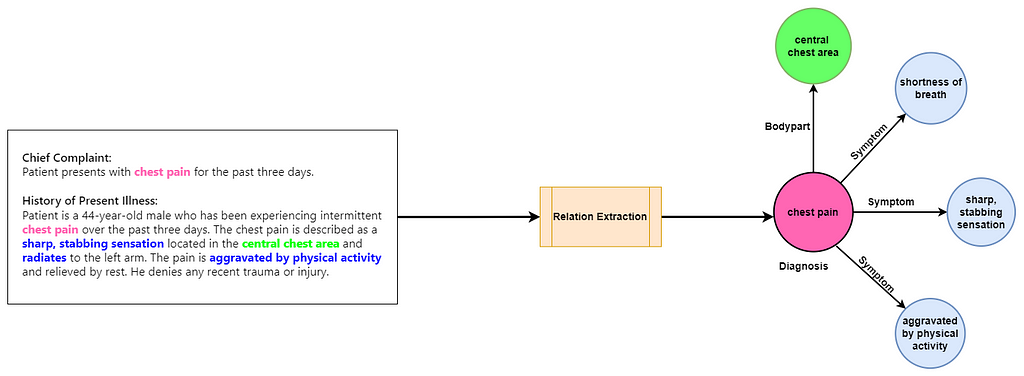
How can LLMs help in automated ICD coding?
When addressing the challenges of automated ICD coding, Large Language Models (LLMs) are well-positioned to address these problems, particularly due to their adaptability to new labels and their ability to manage complex information extraction tasks. However the point here is not to argue that LLMs are the best solution for automated ICD coding, or that these are problems that only LLMs can solve. Rather, by establishing some of the main challenges than an automated ICD coding system must overcome, I analyze how best the abilities of LLMs can be leveraged to solve them.
Adapting to New and Rare ICD Codes:
LLMs demonstrate robust zero-shot and few-shot learning capabilities, allowing them to adapt to new tasks with minimal examples and instructions provided in the prompt. Retrieval-Augmented Generation (RAG) is another paradigm that enables LLMs to access more contextual information to adapt to new tasks without fine-tuning. This is particularly useful for adapting LLMs to new and/or rare ICD codes, which may not be frequently represented in training datasets, from just a few descriptions or examples of usage.
Contextualizing Information:
LLMs are found to be effective at zero-shot relation extraction in the clinical domain [3] [4]. Zero-shot relation extraction allows LLMs to identify and categorize relationships in text without prior specific training on those relationships. This allows for better contextualization of the diagnosis in the medical coding to fetch more precise ICD codes.
Exploring the paper “Automated clinical coding using off-the-shelf large language models”:
While exploring recent works that applied LLMs towards ICD coding, I came across a very interesting paper that leveraged LLMs for ICD coding without any specific fine-tuning. The authors came up with a method which they termed LLM-guided tree-search [5].
How does it work?
The ICD terminology is a hierarchical, tree-like structure. Each ICD code exists within this hierarchical structure where parent codes cover more general conditions, and child codes detail specific diseases. Traversing the ICD tree leads to more specific and fine-grained diagnosis codes.
In LLM-guided tree search, the search begins at the root and uses the LLM to select branches for exploration, continuing iteratively until all paths are exhausted. Practically, this process is implemented by providing the descriptions of all codes at any given level of the tree, along with the medical note, as a prompt to the LLM and asking it to identify the relevant codes for the medical note. The codes selected by the LLM in each instance are then further traversed and explored. This method identifies the most pertinent ICD codes, which are subsequently assigned as predicted labels for the clinical note.
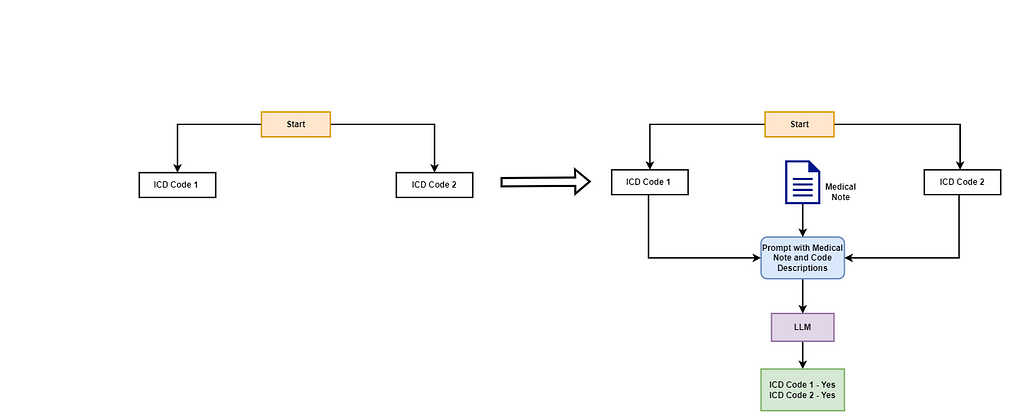
Let’s clarify this with an example. Imagine a tree with two root nodes: ICD Code 1 and ICD Code 2. Each node has a plain-text description characterizing the code. In the initial stage, the LLM is given the medical note along with the descriptions of the codes and asked to identify the codes pertinent to the medical note.

In this scenario, the LLM identifies both ICD Code 1 and ICD Code 2 as relevant to the medical note. The algorithm then examines the child nodes of each code. Each parent code has two child nodes representing more specific ICD codes. Starting with ICD Code 1, the LLM uses the descriptions of ICD Code 1.1 and ICD Code 1.2 along with the medical note to determine the relevant codes. The LLM concludes that ICD Code 1.1 is relevant, while ICD Code 1.2 is not. Since ICD Code 1.1 has no further child nodes, the algorithm checks if it is an assignable code and assigns it to the document. Next, the algorithm evaluates the child nodes of ICD Code 2. Invoking the LLM again, it determines that only ICD Code 2.1 is relevant. This is a simplified example; in reality, the ICD tree is extensive and deeper, meaning the algorithm will continue to traverse the children of each relevant node until it reaches the end of the tree or exhausts valid traversals.
Highlights
- This method does not require any fine-tuning of the LLM; it leverages the LLM’s ability to contextually understand the medical note and dynamically identify the relevant ICD codes based on the provided descriptions.
- Furthermore, this paper shows that LLMs can effectively adapt to a large output space when given relevant information in the prompt, outperforming PLM-ICD [6] on rare codes in terms of macro-average metrics.
- This technique also outperforms the baseline of directly asking the LLM to predict the ICD codes for a medical note based on its parametric knowledge. This highlights the potential in integrating LLMs with tools or external knowledge for solving clinical coding tasks.
Drawbacks
- The algorithm invokes the LLM at every level in the tree. That leads to a high number of LLM invocations as you traverse the tree, compounded by the vastness of the ICD tree. This leads to high latency and costs in processing a single document.
- As the authors also note in the paper, in order to correctly predict a relevant code, the LLM must correctly identify its parent nodes at all levels. Even if a mistake is made at one level, the LLM will be unable to reach the final relevant code.
- The authors were unable to evaluate their method using datasets like MIMIC-III due to limitations that prohibit the transfer of data to external services such as OpenAI’s GPT endpoints. Instead, they evaluated the method using the test set of the CodiEsp dataset [7,8], which includes 250 medical notes. The small size of this dataset suggests that the method’s effectiveness on larger clinical datasets is yet to be established.
Implementing the technique described in the paper
All code and resources related to this article are made available at this link with a mirror of the repo available in my original blog-related repository. I wish to stress that my reimplementation is not exactly identical to the paper and differs in subtle ways that I’ve documented in the original repository. I’ve tried to replicate the prompts used for invoking GPT-3.5 and Llama-70B based on the details in the original paper. For translating the datasets from Spanish to English, I created my own prompt for doing that, as the details were not accessible in the paper.
Let’s implement the technique to better understand how it works. As mentioned, the paper uses the CodiEsp test set for its evaluation. This dataset consists of Spanish medical notes along with their ICD codes. Although the dataset includes an English translated version, the authors note that they translated the Spanish medical notes into English using GPT-3.5, which they claim provided a modest performance improvement over using the pre-translated version. We replicate this functionality and translate the notes into English.
def construct_translation_prompt(medical_note):
"""
Construct a prompt template for translating spanish medical notes to english.
Args:
medical_note (str): The medical case note.
Returns:
str: A structured template ready to be used as input for a language model.
"""
translation_prompt = """You are an expert Spanish-to-English translator. You are provided with a clinical note written in Spanish.
You must translate the note into English. You must ensure that you properly translate the medical and technical terms from Spanish to English without any mistakes.
Spanish Medical Note:
{medical_note}"""
return translation_prompt.format(medical_note = medical_note)
Now that we have the evaluation corpus ready, let’s implement the core logic for the tree-search algorithm. We define the functionality in get_icd_codes, which accepts the medical note to process, the model name, and the temperature setting. The model name must be either “gpt-3.5-turbo-0613” for GPT-3.5 or “meta-llama/Llama-2–70b-chat-hf” for Llama-2 70B Chat. This specification determines the LLM that the tree-search algorithm will invoke during its processing.
Evaluating GPT-4 is possible using the same code-base by providing the appropriate model name, but we choose to skip it as it is quite time-consuming.
def get_icd_codes(medical_note, model_name="gpt-3.5-turbo-0613", temperature=0.0):
"""
Identifies relevant ICD-10 codes for a given medical note by querying a language model.
This function implements the tree-search algorithm for ICD coding described in https://openreview.net/forum?id=mqnR8rGWkn.
Args:
medical_note (str): The medical note for which ICD-10 codes are to be identified.
model_name (str): The identifier for the language model used in the API (default is 'gpt-3.5-turbo-0613').
Returns:
list of str: A list of confirmed ICD-10 codes that are relevant to the medical note.
"""
assigned_codes = []
candidate_codes = [x.name for x in CHAPTER_LIST]
parent_codes = []
prompt_count = 0
while prompt_count < 50:
code_descriptions = {}
for x in candidate_codes:
description, code = get_name_and_description(x, model_name)
code_descriptions[description] = code
prompt = build_zero_shot_prompt(medical_note, list(code_descriptions.keys()), model_name=model_name)
lm_response = get_response(prompt, model_name, temperature=temperature, max_tokens=500)
predicted_codes = parse_outputs(lm_response, code_descriptions, model_name=model_name)
for code in predicted_codes:
if cm.is_leaf(code["code"]):
assigned_codes.append(code["code"])
else:
parent_codes.append(code)
if len(parent_codes) > 0:
parent_code = parent_codes.pop(0)
candidate_codes = cm.get_children(parent_code["code"])
else:
break
prompt_count += 1
return assigned_codes
Similar to the paper, we use the simple_icd_10_cm library, which provides access to the ICD-10 tree. This allows us to traverse the tree, access the descriptions for each code, and identify valid codes. First, we get the nodes at the first level of the tree.
import simple_icd_10_cm as cm
def get_name_and_description(code, model_name):
"""
Retrieve the name and description of an ICD-10 code.
Args:
code (str): The ICD-10 code.
Returns:
tuple: A tuple containing the formatted description and the name of the code.
"""
full_data = cm.get_full_data(code).split("n")
return format_code_descriptions(full_data[3], model_name), full_data[1]
Inside the loop, we obtain the descriptions corresponding to each of the nodes. Now, we need to construct the prompt for the LLM based on the medical note and the code descriptions. We create the prompts for GPT-3.5 and Llama-2 based on the details provided in the paper.
prompt_template_dict = {"gpt-3.5-turbo-0613" : """[Case note]:
{note}
[Example]:
<example prompt>
Gastro-esophageal reflux disease
Enteropotosis
<response>
Gastro-esophageal reflux disease: Yes, Patient was prescribed omeprazole.
Enteropotosis: No.
[Task]:
Consider each of the following ICD-10 code descriptions and evaluate if there are any related mentions in the case note.
Follow the format in the example precisely.
{code_descriptions}""",
"meta-llama/Llama-2-70b-chat-hf": """[Case note]:
{note}
[Example]:
<code descriptions>
* Gastro-esophageal reflux disease
* Enteroptosis
* Acute Nasopharyngitis [Common Cold]
</code descriptions>
<response>
* Gastro-esophageal reflux disease: Yes, Patient was prescribed omeprazole.
* Enteroptosis: No.
* Acute Nasopharyngitis [Common Cold]: No.
</response>
[Task]:
Follow the format in the example response exactly, including the entire description before your (Yes|No) judgement, followed by a newline.
Consider each of the following ICD-10 code descriptions and evaluate if there are any related mentions in the Case note.
{code_descriptions}"""
}
We now construct the prompt based on the medical note and code descriptions. An advantage for us, in terms of prompting and coding, is that we can use the same openai library to interact with both GPT-3.5 and Llama 2, provided that Llama-2 is deployed using deepinfra, which also supports the openai format for sending requests to the LLM.
def construct_prompt_template(case_note, code_descriptions, model_name):
"""
Construct a prompt template for evaluating ICD-10 code descriptions against a given case note.
Args:
case_note (str): The medical case note.
code_descriptions (str): The ICD-10 code descriptions formatted as a single string.
Returns:
str: A structured template ready to be used as input for a language model.
"""
template = prompt_template_dict[model_name]
return template.format(note=case_note, code_descriptions=code_descriptions)
def build_zero_shot_prompt(input_note, descriptions, model_name, system_prompt=""):
"""
Build a zero-shot classification prompt with system and user roles for a language model.
Args:
input_note (str): The input note or query.
descriptions (list of str): List of ICD-10 code descriptions.
system_prompt (str): Optional initial system prompt or instruction.
Returns:
list of dict: A structured list of dictionaries defining the role and content of each message.
"""
if model_name == "meta-llama/Llama-2-70b-chat-hf":
code_descriptions = "n".join(["* " + x for x in descriptions])
else:
code_descriptions = "n".join(descriptions)
input_prompt = construct_prompt_template(input_note, code_descriptions, model_name)
return [{"role": "system", "content": system_prompt}, {"role": "user", "content": input_prompt}]
Having constructed the prompts, we now invoke the LLM to obtain the response:
def get_response(messages, model_name, temperature=0.0, max_tokens=500):
"""
Obtain responses from a specified model via the chat-completions API.
Args:
messages (list of dict): List of messages structured for API input.
model_name (str): Identifier for the model to query.
temperature (float): Controls randomness of response, where 0 is deterministic.
max_tokens (int): Limit on the number of tokens in the response.
Returns:
str: The content of the response message from the model.
"""
response = client.chat.completions.create(
model=model_name,
messages=messages,
temperature=temperature,
max_tokens=max_tokens
)
return response.choices[0].message.content
Great, we’ve obtained the output! From the response, we now parse each code description to identify the nodes that the LLM has deemed relevant for further traversal, as well as those nodes the LLM has rejected. We break the output response into new lines and split each response to identify the prediction of the LLM for each code description.
def remove_noisy_prefix(text):
# Removing numbers or letters followed by a dot and optional space at the beginning of the string
cleaned_text = text.replace("* ", "").strip()
cleaned_text = re.sub(r"^s*w+.s*", "", cleaned_text)
return cleaned_text.strip()
def parse_outputs(output, code_description_map, model_name):
"""
Parse model outputs to confirm ICD-10 codes based on a given description map.
Args:
output (str): The model output containing confirmations.
code_description_map (dict): Mapping of descriptions to ICD-10 codes.
Returns:
list of dict: A list of confirmed codes and their descriptions.
"""
confirmed_codes = []
split_outputs = [x for x in output.split("n") if x]
for item in split_outputs:
try:
code_description, confirmation = item.split(":", 1)
if model_name == "meta-llama/Llama-2-70b-chat-hf":
code_description = remove_noisy_prefix(code_description)
if confirmation.lower().strip().startswith("yes"):
try:
code = code_description_map[code_description]
confirmed_codes.append({"code": code, "description": code_description})
except Exception as e:
print(str(e) + " Here")
continue
except:
continue
return confirmed_codes
Let’s look at the remainder of the loop now. So far, we have constructed the prompt, obtained the response from the LLM, and parsed the output to identify the codes deemed relevant by the LLM.
while prompt_count < 50:
code_descriptions = {}
for x in candidate_codes:
description, code = get_name_and_description(x, model_name)
code_descriptions[description] = code
prompt = build_zero_shot_prompt(medical_note, list(code_descriptions.keys()), model_name=model_name)
lm_response = get_response(prompt, model_name, temperature=temperature, max_tokens=500)
predicted_codes = parse_outputs(lm_response, code_descriptions, model_name=model_name)
for code in predicted_codes:
if cm.is_leaf(code["code"]):
assigned_codes.append(code["code"])
else:
parent_codes.append(code)
if len(parent_codes) > 0:
parent_code = parent_codes.pop(0)
candidate_codes = cm.get_children(parent_code["code"])
else:
break
prompt_count += 1
Now we iterate through the predicted codes and check if each code is a “leaf” code, which essentially ensures that the code is a valid and assignable ICD code. If the predicted code is valid, we consider it as a prediction by the LLM for that medical note. If not, we add it to our parent codes and obtain the children nodes to further traverse the ICD tree. We break out of the loop if there are no more parent codes to further traverse.
In theory, the number of LLM invocations per medical note can be arbitrarily high, leading to increased latency if the algorithm traverses many nodes. The authors enforce a maximum of 50 prompts/LLM invocations per medical note to terminate the processing, a limit we also adopt in our implementation.
Results
We can now evaluate the results of the tree-search algorithm using GPT-3.5 and Llama-2 as the LLMs. We assess the performance of the algorithm in terms of micro-average and macro-average precision, recall, and F1-score.

While the implementation’s results are roughly in the ball-park of the reported scores in the paper, there are some note-worthy differences.
- In this implementation, GPT-3.5’s micro-average metrics slightly exceed the reported figures, while the macro-average metrics fall a bit short of the reported values.
- Similarly, Llama-70B’s micro-average metrics either match or slightly exceed the reported figures, but the macro-average metrics are lower than the reported values.
As mentioned earlier, this implementation differs from the paper in a few minor ways, all of which impact the final performance. Please refer to the linked repository for a more detailed discussion of how this implementation differs from the original paper.
Conclusion
Understanding and implementing this method was quite insightful for me in many ways. It allowed me to develop a more nuanced understanding of the strengths and weaknesses of Large Language Models (LLMs) in the clinical coding case. Specifically, it became evident that when LLMs have dynamic access to pertinent information about the codes, they can effectively comprehend the clinical context and accurately identify the relevant codes.
It would be interesting to explore whether utilizing LLMs as agents for clinical coding could further improve performance. Given the abundance of external knowledge sources for biomedical and clinical texts in the form of papers or knowledge graphs, LLM agents could potentially be used in workflows that analyze medical documents at a finer granularity. They could also invoke tools that allow them to refer to external knowledge on the fly if required, to arrive at the final code.
Acknowledgement
Huge thanks to Joseph, the lead author of this paper, for clarifying my doubts regarding the evaluation of this method!
References:
[1] https://www.who.int/standards/classifications/classification-of-diseases
[2] Johnson, A. E., Pollard, T. J., Shen, L., Lehman, L. W. H., Feng, M., Ghassemi, M., … & Mark, R. G. (2016). MIMIC-III, a freely accessible critical care database Sci. Data, 3(1), 1.
[3] Agrawal, M., Hegselmann, S., Lang, H., Kim, Y., & Sontag, D. (2022). Large language models are few-shot clinical information extractors. arXiv preprint arXiv:2205.12689.
[4] Zhou, H., Li, M., Xiao, Y., Yang, H., & Zhang, R. (2023). LLM Instruction-Example Adaptive Prompting (LEAP) Framework for Clinical Relation Extraction. medRxiv : the preprint server for health sciences, 2023.12.15.23300059. https://doi.org/10.1101/2023.12.15.23300059
[5] Boyle, J. S., Kascenas, A., Lok, P., Liakata, M., & O’Neil, A. Q. (2023, October). Automated clinical coding using off-the-shelf large language models. In Deep Generative Models for Health Workshop NeurIPS 2023.
[6] Huang, C. W., Tsai, S. C., & Chen, Y. N. (2022). PLM-ICD: automatic ICD coding with pretrained language models. arXiv preprint arXiv:2207.05289.
[7] Miranda-Escalada, A., Gonzalez-Agirre, A., Armengol-Estapé, J., & Krallinger, M. (2020). Overview of Automatic Clinical Coding: Annotations, Guidelines, and Solutions for non-English Clinical Cases at CodiEsp Track of CLEF eHealth 2020. CLEF (Working Notes), 2020.
[8] Miranda-Escalada, A., Gonzalez-Agirre, A., & Krallinger, M. (2020). CodiEsp corpus: gold standard Spanish clinical cases coded in ICD10 (CIE10) — eHealth CLEF2020 (1.4) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.3837305 (CC BY 4.0)
Exploring LLMs for ICD Coding — Part 1 was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
Originally appeared here:
Exploring LLMs for ICD Coding — Part 1
Go Here to Read this Fast! Exploring LLMs for ICD Coding — Part 1