How can an LLM be applied effectively for biomedical entity linking?

Biomedical text is a catch-all term that broadly encompasses documents such as research articles, clinical trial reports, and patient records, serving as rich repositories of information about various biological, medical, and scientific concepts. Research papers in the biomedical field present novel breakthroughs in areas like drug discovery, drug side effects, and new disease treatments. Clinical trial reports offer in-depth details on the safety, efficacy, and side effects of new medications or treatments. Meanwhile, patient records contain comprehensive medical histories, diagnoses, treatment plans, and outcomes recorded by physicians and healthcare professionals.
Mining these texts allows practitioners to extract valuable insights, which can be beneficial for various downstream tasks. You could mine text to identify adverse drug reaction extractions, build automated medical coding algorithms or build information retrieval or question-answering systems that can help extract information from vast research corpora. However, one issue affecting biomedical document processing is the often unstructured nature of the text. For example, researchers might use different terms to refer to the same concept. What one researcher calls a “heart attack” might be referred to as a “myocardial infarction” by another. Similarly, in drug-related documentation, technical and common names may be used interchangeably. For instance, “Acetaminophen” is the technical name of a drug, while “Paracetamol” is its more common counterpart. The prevalence of abbreviations also adds another layer of complexity; for instance, “Nitric Oxide” might be referred to as “NO” in another context. Despite these varying terms referring to the same concept, these variations make it difficult for a layman or a text-processing algorithm to determine whether they refer to the same concept. Thus, Entity Linking becomes crucial in this situation.
Table of Contents:
- What is Entity Linking?
- Where do LLMs come in here?
- Experimental Setup
- Processing the Dataset
- Zero-Shot Entity Linking using the LLM
- LLM with Retrieval Augmented Generation for Entity Linking
- Zero-Shot Entity Extraction with the LLM and an External KB Linker
- Fine-tuned Entity Extraction with the LLM and an External KB Linker
- Benchmarking Scispacy
- Takeaways
- Limitations
- References
What is Entity Linking?
When text is unstructured, accurately identifying and standardizing medical concepts becomes crucial. To achieve this, medical terminology systems such as Unified Medical Language System (UMLS) [1], Systematized Medical Nomenclature for Medicine–Clinical Terminology (SNOMED-CT) [2], and Medical Subject Headings (MeSH) [3] play an essential role. These systems provide a comprehensive and standardized set of medical concepts, each uniquely identified by an alphanumeric code.
Entity linking involves recognizing and extracting entities within the text and mapping them to standardized concepts in a large terminology. In this context, a Knowledge Base (KB) refers to a detailed database containing standardized information and concepts related to the terminology, such as medical terms, diseases, and drugs. Typically, a KB is expert-curated and designed, containing detailed information about the concepts, including variations of the terms that could be used to refer to the concept, or how it is related to other concepts.

Entity recognition entails extracting words or phrases that are significant in the context of our task. In this context, it usually refers to extraction of biomedical terms such as drugs, diseases etc. Typically, lookup-based methods or machine learning/deep learning-based systems are often used for entity recognition. Linking the entities to a KB usually involves a retriever system that indexes the KB. This system takes each extracted entity from the previous step and retrieves likely identifiers from the KB. The retriever here is also an abstraction, which may be sparse (BM-25), dense (embedding-based), or even a generative system (like a Large Language Model, (LLM)) that has encoded the KB in its parameters.
Where do LLMs come in here?
I’ve been curious for a while about the best ways to integrate LLMs into biomedical and clinical text-processing pipelines. Given that Entity Linking is an important part of such pipelines, I decided to explore how best LLMs can be utilized for this task. Specifically I investigated the following setups:
- Zero-Shot Entity Linking with an LLM: Leveraging an LLM to directly identify all entities and concept IDs from input biomedical texts without any fine-tuning
- LLM with Retrieval Augmented Generation (RAG): Utilizing the LLM within a RAG framework by injecting information about relevant concept IDs in the prompt to identify the relevant concept IDs.
- Zero-Shot Entity Extraction with LLM with an External KB Linker: Employing the LLM for zero-shot entity extraction from biomedical texts, with an external linker/retriever for mapping the entities to concept IDs.
- Fine-tuned Entity Extraction with an External KB Linker: Finetuning the LLM first on the entity extraction task, and using it as an entity extractor with an external linker/retriever for mapping the entities to concept IDs.
- Comparison with an existing pipeline: How do these methods fare comparted to Scispacy, a commonly used library for biomedical text processing?
Experimental Setup
All code and resources related to this article are made available at this Github repository, under the entity_linking folder. Feel free to pull the repository and run the notebooks directly to run these experiments. Please let me know if you have any feedback or observations or if you notice any mistakes!
To conduct these experiments, we utilize the Mistral-7B Instruct model [9] as our Large Language Model (LLM). For the medical terminology to link entities against, we utilize the MeSH terminology. To quote the National Library of Medicine website:
“The Medical Subject Headings (MeSH) thesaurus is a controlled and hierarchically-organized vocabulary produced by the National Library of Medicine. It is used for indexing, cataloging, and searching of biomedical and health-related information.”
We utilize the BioCreative-V-CDR-Corpus [4,5,6,7,8] for evaluation. This dataset contains annotations of disease and chemical entities, along with their corresponding MeSH IDs. For evaluation purposes, we randomly sample 100 data points from the test set. We used a version of the MeSH KB provided by Scispacy [10,11], which contains information about the MeSH identifiers, such as definitions and entities corresponding to each ID.
For performance evaluation, we calculate two metrics. The first metric relates to the entity extraction performance. The original dataset contains all mentions of entities in the text, annotated at the substring level. A strict evaluation would check if the algorithm has outputted all occurrences of all entities. However, we simplify this process for easier evaluation; we lower-case and de-duplicate the entities in the ground truth. We then calculated the Precision, Recall and F1 score for each instance and calculate the macro-average for each metric.
Suppose you have a set of actual entities, ground_truth, and a set of entities predicted by a model, pred for each input text. The true positives TP can be determined by identifying the common elements between pred and ground_truth, essentially by calculating the intersection of these two sets.
For each input, we can then calculate:
precision = len(TP)/ len(pred) ,
recall = len(TP) / len(ground_truth) and
f1 = 2 * precision * recall / (precision + recall)
and finally calculate the macro-average for each metric by summing them all up and dividing by the number of datapoints in our test set.
For evaluating the overall entity linking performance, we again calculate the same metrics. In this case, for each input datapoint, we have a set of tuples, where each tuple is a (entity, mesh_id) pair. The metrics are otherwise calculated the same way.
Processing the Dataset
Right, let’s kick off things by first defining some helper functions for processing our dataset.
def parse_dataset(file_path):
"""
Parse the BioCreative Dataset.
Args:
- file_path (str): Path to the file containing the documents.
Returns:
- list of dict: A list where each element is a dictionary representing a document.
"""
documents = []
current_doc = None
with open(file_path, 'r', encoding='utf-8') as file:
for line in file:
line = line.strip()
if not line:
continue
if "|t|" in line:
if current_doc:
documents.append(current_doc)
id_, title = line.split("|t|", 1)
current_doc = {'id': id_, 'title': title, 'abstract': '', 'annotations': []}
elif "|a|" in line:
_, abstract = line.split("|a|", 1)
current_doc['abstract'] = abstract
else:
parts = line.split("t")
if parts[1] == "CID":
continue
annotation = {
'text': parts[3],
'type': parts[4],
'identifier': parts[5]
}
current_doc['annotations'].append(annotation)
if current_doc:
documents.append(current_doc)
return documents
def deduplicate_annotations(documents):
"""
Filter documents to ensure annotation consistency.
Args:
- documents (list of dict): The list of documents to be checked.
"""
for doc in documents:
doc["annotations"] = remove_duplicates(doc["annotations"])
def remove_duplicates(dict_list):
"""
Remove duplicate dictionaries from a list of dictionaries.
Args:
- dict_list (list of dict): A list of dictionaries from which duplicates are to be removed.
Returns:
- list of dict: A list of dictionaries after removing duplicates.
"""
unique_dicts = []
seen = set()
for d in dict_list:
dict_tuple = tuple(sorted(d.items()))
if dict_tuple not in seen:
seen.add(dict_tuple)
unique_dicts.append(d)
return unique_dicts
We first parse the dataset from the text files provided in the original dataset. The original dataset includes the title, abstract, and all entities annotated with their entity type (Disease or Chemical), their substring indices indicating their exact location in the text, along with their MeSH IDs. While processing our dataset, we make a few simplifications. We disregard the substring indices and the entity type. Moreover, we de-duplicate annotations that share the same entity name and MeSH ID. At this stage, we only de-duplicate in a case-sensitive manner, meaning if the same entity appears in both lower and upper case across the document, we retain both instances in our processing so far.
Zero-Shot Entity Linking using the LLM
First, we aim to determine whether the LLM already possesses an understanding of MeSH terminology due to its pre-training, and if it can function as a zero-shot entity linker. By zero-shot, we mean the LLM’s capability to directly link entities to their MeSH IDs from biomedical text based on its intrinsic knowledge, without depending on an external KB linker. This hypothesis is not entirely unrealistic, considering the availability of information about MeSH online, which makes it possible that the model might have encountered MeSH-related information during its pre-training phase. However, even if the LLM was trained with such information, it is unlikely that this alone would enable the model to perform zero-shot entity linking effectively, due to the complexity of biomedical terminology and the precision required for accurate entity linking.
To evaluate this, we provide the input text to the LLM and directly prompt it to predict the entities and corresponding MeSH IDs. Additionally, we create a few-shot prompt by sampling three data points from the training dataset. It is important to clarify the distinction in the use of “zero-shot” and “few-shot” here: “zero-shot” refers to the LLM as a whole performing entity linking without prior specific training on this task, while “few-shot” refers to the prompting strategy employed in this context.

To calculate our metrics, we define functions for evaluating the performance:
def calculate_entity_metrics(gt, pred):
"""
Calculate precision, recall, and F1-score for entity recognition.
Args:
- gt (list of dict): A list of dictionaries representing the ground truth entities.
Each dictionary should have a key "text" with the entity text.
- pred (list of dict): A list of dictionaries representing the predicted entities.
Similar to `gt`, each dictionary should have a key "text".
Returns:
tuple: A tuple containing precision, recall, and F1-score (in that order).
"""
ground_truth_set = set([x["text"].lower() for x in gt])
predicted_set = set([x["text"].lower() for x in pred])
# True positives are predicted items that are in the ground truth
true_positives = len(predicted_set.intersection(ground_truth_set))
# Precision calculation
if len(predicted_set) == 0:
precision = 0
else:
precision = true_positives / len(predicted_set)
# Recall calculation
if len(ground_truth_set) == 0:
recall = 0
else:
recall = true_positives / len(ground_truth_set)
# F1-score calculation
if precision + recall == 0:
f1_score = 0
else:
f1_score = 2 * (precision * recall) / (precision + recall)
return precision, recall, f1_score
def calculate_mesh_metrics(gt, pred):
"""
Calculate precision, recall, and F1-score for matching MeSH (Medical Subject Headings) codes.
Args:
- gt (list of dict): Ground truth data
- pred (list of dict): Predicted data
Returns:
tuple: A tuple containing precision, recall, and F1-score (in that order).
"""
ground_truth = []
for item in gt:
mesh_codes = item["identifier"]
if mesh_codes == "-1":
mesh_codes = "None"
mesh_codes_split = mesh_codes.split("|")
for elem in mesh_codes_split:
combined_elem = {"entity": item["text"].lower(), "identifier": elem}
if combined_elem not in ground_truth:
ground_truth.append(combined_elem)
predicted = []
for item in pred:
mesh_codes = item["identifier"]
mesh_codes_split = mesh_codes.strip().split("|")
for elem in mesh_codes_split:
combined_elem = {"entity": item["text"].lower(), "identifier": elem}
if combined_elem not in predicted:
predicted.append(combined_elem)
# True positives are predicted items that are in the ground truth
true_positives = len([x for x in predicted if x in ground_truth])
# Precision calculation
if len(predicted) == 0:
precision = 0
else:
precision = true_positives / len(predicted)
# Recall calculation
if len(ground_truth) == 0:
recall = 0
else:
recall = true_positives / len(ground_truth)
# F1-score calculation
if precision + recall == 0:
f1_score = 0
else:
f1_score = 2 * (precision * recall) / (precision + recall)
return precision, recall, f1_score
Let’s now run the model and get our predictions:
model = AutoModelForCausalLM.from_pretrained("mistralai/Mistral-7B-Instruct-v0.2", torch_dtype=torch.bfloat16).cuda()
tokenizer = AutoTokenizer.from_pretrained("mistralai/Mistral-7B-Instruct-v0.2")
model.eval()
mistral_few_shot_answers = []
for item in tqdm(test_set_subsample):
few_shot_prompt_messages = build_few_shot_prompt(SYSTEM_PROMPT, item, few_shot_example)
input_ids = tokenizer.apply_chat_template(few_shot_prompt_messages, tokenize=True, return_tensors = "pt").cuda()
outputs = model.generate(input_ids = input_ids, max_new_tokens=200, do_sample=False)
# https://github.com/huggingface/transformers/issues/17117#issuecomment-1124497554
gen_text = tokenizer.batch_decode(outputs.detach().cpu().numpy()[:, input_ids.shape[1]:], skip_special_tokens=True)[0]
mistral_few_shot_answers.append(parse_answer(gen_text.strip()))
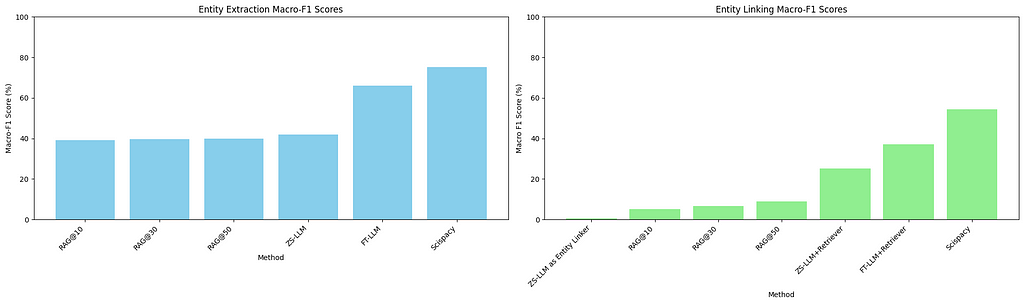
At the entity extraction level, the LLM performs quite well, considering it has not been explicitly fine-tuned for this task. However, its performance as a zero-shot linker is quite poor, with an overall performance of less than 1%. This outcome is intuitive, though, because the output space for MeSH labels is vast, and it is a hard task to exactly map entities to a specific MeSH ID.
LLM with Retrieval Augmented Generation for Entity Linking
Retrieval Augmented Generation (RAG) [12] refers to a framework that combines LLMs with an external KB equipped with a querying function, such as a retriever/linker. For each incoming query, the system first retrieves knowledge relevant to the query from the KB using the querying function. It then combines the retrieved knowledge and the query, providing this combined prompt to the LLM to perform the task. This approach is based on the understanding that LLMs may not have all the necessary knowledge or information to answer an incoming query effectively. Thus, knowledge is injected into the model by querying an external knowledge source.
Using a RAG framework can offer several advantages:
- An existing LLM can be utilized for a new domain or task without the need for domain-specific fine-tuning, as the relevant information can be queried and provided to the model through a prompt.
- LLMs can sometimes provide incorrect answers (hallucinate) when responding to queries. Employing RAG with LLMs can significantly reduce such hallucinations, as the answers provided by the LLM are more likely to be grounded in facts due to the knowledge supplied to it.
Considering that the LLM lacks specific knowledge of MeSH terminologies, we investigate whether a RAG setup could enhance performance. In this approach, for each input paragraph, we utilize a BM-25 retriever to query the KB. For each MeSH ID, we have access to a general description of the ID and the entity names associated with it. After retrieval, we inject this information to the model through the prompt for entity linking.
To investigate the effect of the number of retrieved IDs provided as context to the model on the entity linking process, we run this setup by providing top 10, 30 and 50 documents to the model and quantify its performance on entity extraction and MeSH concept identification.
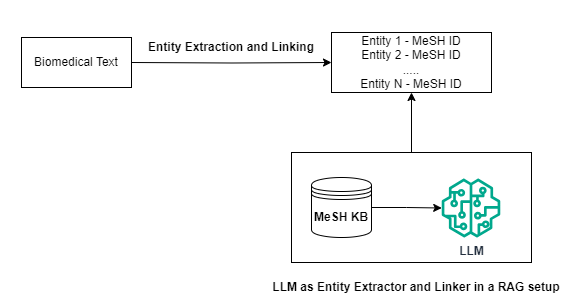
Let’s first define our BM-25 Retriever:
from rank_bm25 import BM25Okapi
from typing import List, Tuple, Dict
from nltk.tokenize import word_tokenize
from tqdm import tqdm
class BM25Retriever:
"""
A class for retrieving documents using the BM25 algorithm.
Attributes:
index (List[int, str]): A dictionary with document IDs as keys and document texts as values.
tokenized_docs (List[List[str]]): Tokenized version of the documents in `processed_index`.
bm25 (BM25Okapi): An instance of the BM25Okapi model from the rank_bm25 package.
"""
def __init__(self, docs_with_ids: Dict[int, str]):
"""
Initializes the BM25Retriever with a dictionary of documents.
Args:
docs_with_ids (List[List[str, str]]): A dictionary with document IDs as keys and document texts as values.
"""
self.index = docs_with_ids
self.tokenized_docs = self._tokenize_docs([x[1] for x in self.index])
self.bm25 = BM25Okapi(self.tokenized_docs)
def _tokenize_docs(self, docs: List[str]) -> List[List[str]]:
"""
Tokenizes the documents using NLTK's word_tokenize.
Args:
docs (List[str]): A list of documents to be tokenized.
Returns:
List[List[str]]: A list of tokenized documents.
"""
return [word_tokenize(doc.lower()) for doc in docs]
def query(self, query: str, top_n: int = 10) -> List[Tuple[int, float]]:
"""
Queries the BM25 model and retrieves the top N documents with their scores.
Args:
query (str): The query string.
top_n (int): The number of top documents to retrieve.
Returns:
List[Tuple[int, float]]: A list of tuples, each containing a document ID and its BM25 score.
"""
tokenized_query = word_tokenize(query.lower())
scores = self.bm25.get_scores(tokenized_query)
doc_scores_with_ids = [(doc_id, scores[i]) for i, (doc_id, _) in enumerate(self.index)]
top_doc_ids_and_scores = sorted(doc_scores_with_ids, key=lambda x: x[1], reverse=True)[:top_n]
return [x[0] for x in top_doc_ids_and_scores]
We now process our KB file and create a BM-25 retriever instance that indexes it. While indexing the KB, we index each ID using a concatenation of their description, aliases and canonical name.
def process_index(index):
"""
Processes the initial document index to combine aliases, canonical names, and definitions into a single text index.
Args:
- index (Dict): The MeSH knowledge base
Returns:
List[List[int, str]]: A dictionary with document IDs as keys and combined text indices as values.
"""
processed_index = []
for key, value in tqdm(index.items()):
assert(type(value["aliases"]) != list)
aliases_text = " ".join(value["aliases"].split(","))
text_index = (aliases_text + " " + value.get("canonical_name", "")).strip()
if "definition" in value:
text_index += " " + value["definition"]
processed_index.append([value["concept_id"], text_index])
return processed_index
mesh_data = read_jsonl_file("mesh_2020.jsonl")
process_mesh_kb(mesh_data)
mesh_data_kb = {x["concept_id"]:x for x in mesh_data}
mesh_data_dict = process_index({x["concept_id"]:x for x in mesh_data})
retriever = BM25Retriever(mesh_data_dict)
mistral_rag_answers = {10:[], 30:[], 50:[]}
for k in [10,30,50]:
for item in tqdm(test_set_subsample):
relevant_mesh_ids = retriever.query(item["title"] + " " + item["abstract"], top_n = k)
relevant_contexts = [mesh_data_kb[x] for x in relevant_mesh_ids]
rag_prompt = build_rag_prompt(SYSTEM_RAG_PROMPT, item, relevant_contexts)
input_ids = tokenizer.apply_chat_template(rag_prompt, tokenize=True, return_tensors = "pt").cuda()
outputs = model.generate(input_ids = input_ids, max_new_tokens=200, do_sample=False)
gen_text = tokenizer.batch_decode(outputs.detach().cpu().numpy()[:, input_ids.shape[1]:], skip_special_tokens=True)[0]
mistral_rag_answers[k].append(parse_answer(gen_text.strip()))
entity_scores_at_k = {}
mesh_scores_at_k = {}
for key, value in mistral_rag_answers.items():
entity_scores = [calculate_entity_metrics(gt["annotations"],pred) for gt, pred in zip(test_set_subsample, value)]
macro_precision_entity = sum([x[0] for x in entity_scores]) / len(entity_scores)
macro_recall_entity = sum([x[1] for x in entity_scores]) / len(entity_scores)
macro_f1_entity = sum([x[2] for x in entity_scores]) / len(entity_scores)
entity_scores_at_k[key] = {"macro-precision": macro_precision_entity, "macro-recall": macro_recall_entity, "macro-f1": macro_f1_entity}
mesh_scores = [calculate_mesh_metrics(gt["annotations"],pred) for gt, pred in zip(test_set_subsample, value)]
macro_precision_mesh = sum([x[0] for x in mesh_scores]) / len(mesh_scores)
macro_recall_mesh = sum([x[1] for x in mesh_scores]) / len(mesh_scores)
macro_f1_mesh = sum([x[2] for x in mesh_scores]) / len(mesh_scores)
mesh_scores_at_k[key] = {"macro-precision": macro_precision_mesh, "macro-recall": macro_recall_mesh, "macro-f1": macro_f1_mesh}
In general, the RAG setup improves the overall MeSH Identification process, compared to the original zero-shot setup. But what is the impact of the number of documents provided as information to the model? We plot the scores as a function of the number of retrieved IDs provided to the model as context.
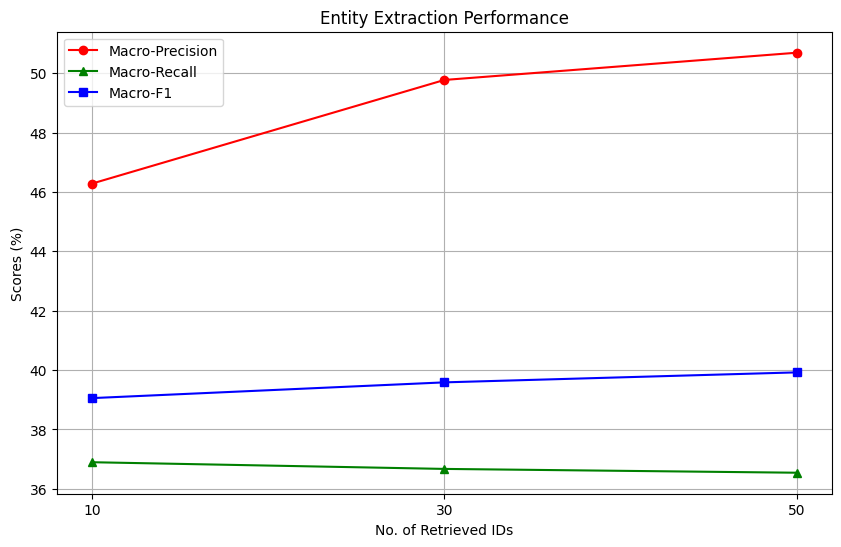
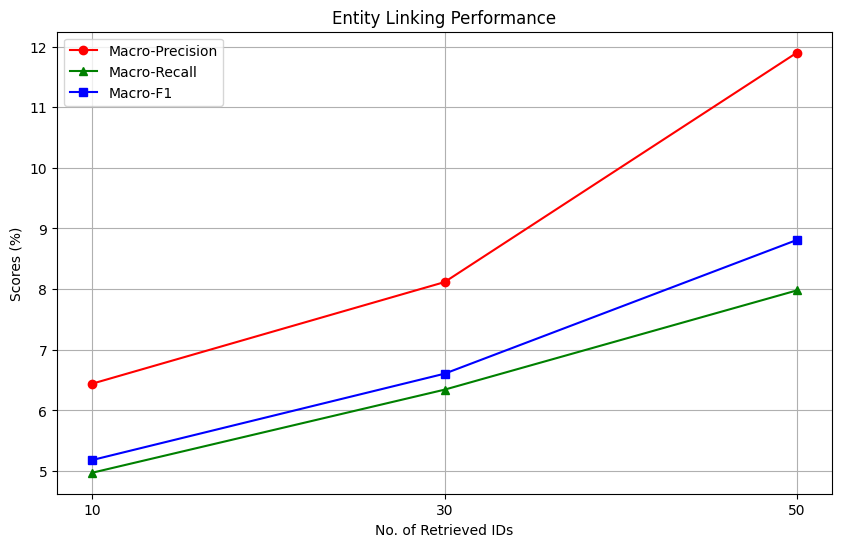
We observe interesting trends while investigating the plots. For entity extraction, an increase in the number of retrieved documents correlates with a sharp increase in macro-precision, reaching a score of slightly higher than 50%. This is nearly 10% higher than the zero-shot entity extraction performance of the model. However, the impact on macro-recall is task-dependent; it remains unchanged for entity extraction but improves for entity linking. Overall, increasing the number of documents provided to the model as context improves all metrics significantly in the MeSH Identification setting, but has mixed gains in the entity extraction setting.
An important limitation to consider in this experiment is the performance of the upstream retriever. If the retriever fails to retrieve relevant documents, the performance of the LLM will suffer as a consequence because the actual answer is not present in the knowledge provided to the model.

To investigate this, we calculated the average % of ground truth MeSH IDs present in the MeSH IDs fetched by the retriever per input text. Our findings show that the BM-25 retriever manages to retrieve only about 12.6% to 17.7% of the relevant MeSH IDs for each input data point on average. The choice of retriever and the way we retrieve is therefore a significant performance bottleneck for the RAG setup and can potentially be optimized for better performance.
Zero-Shot Entity Extraction with the LLM and an External KB Linker
So far, we’ve examined how the LLM performs as a zero-shot entity linker and to what extent RAG can enhance its performance. Though RAG improves performance compared to the zero-shot setup, there are limitations to this approach.
When using LLMs in a RAG setup, we have kept the knowledge component (KB + retriever) upstream of the model until now. The retrieval of knowledge in the RAG setup is coarse, in that we retrieve possible MeSH IDs by querying the retriever using the entire biomedical text. This ensures diversity to a certain extent in the retrieved results, as the fetched results are likely to correspond to different entities in the text, but the results are less likely to be precise. This may not seem like a problem at first, because you can mitigate this to a certain degree by providing more relevant results as context to the model in the RAG setting. However, this has two drawbacks:
- LLMs generally have an upper bound on the context length for processing text. The context length of an LLM roughly refers to the maximum number of tokens the LLM can take into account (the number of tokens in the prompt) before generating new text. This can restrict the amount of knowledge we can provide to the LLM.
- Let’s assume we have an LLM capable of processing long context lengths. We can now retrieve and append more context to the model. Great! However, a longer context length may not necessarily correlate with enhanced RAG abilities for the LLM [13]. Even if you pass a lot of relevant knowledge to the LLM by retrieving more results, this does not guarantee that the LLM will accurately extract the correct answer.
This brings us back to the traditional pipeline of entity linking as described initially. In this setting, the knowledge component is kept downstream to the model, where after entity extraction, the entities are provided to an external retriever for obtaining the relevant MeSH ID. Provided you have a good entity extractor, you can retrieve more precise MeSH IDs.
Earlier, we observed in the fully zero-shot setting that, while the LLM was poor at predicting the MeSH ID, its entity extraction performance was quite decent. We now extract the entities using the Mistral model and provide them to an external retriever for fetching the MeSH IDs.
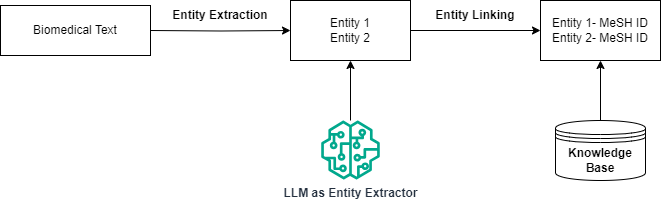
For retrieval here, we again use a BM-25 retriever as our KB linker. However, a small change we make here is to index our IDs based on concatenating their canonical name and aliases. We re-use the entities extracted from the first zero-shot setup for our experiment here. Let’s now evaluate how well this setup performs:
entity_mesh_data_dict = [[x["concept_id"] , " ".join(x["aliases"].split(",")) + " " + x["canonical_name"]] for x in mesh_data]
entity_retriever = BM25Retriever(entity_mesh_data_dict)
parsed_entities_few_shot = [[y["text"] for y in x] for x in mistral_few_shot_answers]
retrieved_answers = []
for item in tqdm(parsed_entities_few_shot):
answer_element = []
for entity in item:
retrieved_mesh_ids = entity_retriever.query(entity, top_n = 1)
answer_element.append({"text": entity, "identifier":retrieved_mesh_ids[0]})
retrieved_answers.append(answer_element)
mesh_scores = [calculate_mesh_metrics(gt["annotations"],pred) for gt, pred in zip(test_set_subsample, retrieved_answers)]
macro_precision_mesh = sum([x[0] for x in mesh_scores]) / len(metric_scores)
macro_recall_mesh = sum([x[1] for x in mesh_scores]) / len(metric_scores)
macro_f1_mesh = sum([x[2] for x in mesh_scores]) / len(metric_scores)
The performance in this setting significantly improves over the RAG setting across all the metrics. We achieve more than 12% improvement in Macro-Precision, 20% improvement in Macro-Recall and 16% improvement in Macro-F1 scores compared to the best RAG setting (retrieval at 50 documents). To stress the point again, this is more akin to the traditional pipeline of entity extraction where you have entity extraction and linking as separate components.
Fine-tuned Entity Extraction with the LLM and an External KB Linker
Until now, we got the best performance by using the LLM as an entity extractor within a larger pipeline. However, we did the entity extraction in a zero-shot manner. Could we achieve further performance gains by fine-tuning the LLM specifically for entity extraction?
For fine-tuning, we utilize the training set from the BioCreative V dataset, which consists of 500 data points. We employ Q-Lora [14] for fine-tuning our LLM, a process that involves quantizing our LLM to 4-bit and freezing it, while fine-tuning a Low-Rank Adapter. This approach is generally parameter and memory efficient, as the Adapter possesses only a fraction of the weights compared to the original LLM, meaning we are fine-tuning significantly fewer weights than if we were to fine-tune the entire LLM. It also enables us to fine-tune our model on a single GPU.
Let’s implement the fine-tuning component. For this part, I referred to and modified Niels Rogge’s notebook on fine-tuning a Mistral Model with Q-Lora, for fine-tuning the model, with the modifications mostly around correctly preparing and processing the dataset.
from datasets import load_dataset
import json
from tqdm import tqdm
from itertools import chain
from datasets import DatasetDict
from transformers import AutoTokenizer, BitsAndBytesConfig
import torch
from trl import SFTTrainer
from peft import LoraConfig
from transformers import TrainingArguments
from helpers import *
def read_jsonl_file(file_path):
"""
Parses a JSONL (JSON Lines) file and returns a list of dictionaries.
Args:
file_path (str): The path to the JSONL file to be read.
Returns:
list of dict: A list where each element is a dictionary representing
a JSON object from the file.
"""
jsonl_lines = []
with open(file_path, 'r', encoding="utf-8") as file:
for line in file:
json_object = json.loads(line)
jsonl_lines.append(json_object)
return jsonl_lines
def convert_to_template(data):
messages = []
messages.append({"role": "user", "content": data["question"]})
messages.append({"role": "assistant", "content": data["answer"]})
return tokenizer.apply_chat_template(messages, tokenize = False)
mesh_dataset = parse_dataset("CDR_TrainingSet.PubTator.txt")
We now load the tokenizer and set the appropriate parameters:
model_id = "mistralai/Mistral-7B-Instruct-v0.2"
tokenizer = AutoTokenizer.from_pretrained(model_id)
# set pad_token_id equal to the eos_token_id if not set
tokenizer.pad_token_id = tokenizer.eos_token_id
tokenizer.padding_side = "right"
# Set reasonable default for models without max length
if tokenizer.model_max_length > 100_000:
tokenizer.model_max_length = 512
Let’s now prepare and format our dataset properly. We define the prompts for our model and format our datasets in the expected chat template.
prepared_dataset = []
system_prompt = "Answer the question factually and precisely."
entity_prompt = "What are the chemical and disease related entities present in this biomedical text?"
prepared_dataset = []
def prepare_instructions(elem):
entities = []
for x in elem["annotations"]:
if x["text"] not in entities:
entities.append(x["text"])
return {"question": system_prompt + "n" + entity_prompt + "n" + elem["title"] + " " + elem["abstract"] , "answer": "The entities are:" + ",".join(entities)}
questions = [prepare_instructions(x) for x in tqdm(mesh_dataset)]
chat_format_questions = [{"text": convert_to_template(x)} for x in tqdm(questions)]
df = pd.DataFrame(chat_format_questions)
train_dataset = Dataset.from_pandas(df)
Let’s now define the appropriate configs for fine-tuning our model. We define the configuration for quantizing the LLM:
quantization_config = BitsAndBytesConfig(
load_in_4bit=True,
bnb_4bit_quant_type="nf4",
bnb_4bit_compute_dtype=torch.bfloat16,
)
device_map = {"": torch.cuda.current_device()} if torch.cuda.is_available() else None
model_kwargs = dict(
torch_dtype=torch.bfloat16,
use_cache=False, # set to False as we're going to use gradient checkpointing
device_map=device_map,
quantization_config=quantization_config,
)
Now, we are all-set to finetune our model:
output_dir = 'entity_finetune'
# based on config
training_args = TrainingArguments(
bf16=True, # specify bf16=True instead when training on GPUs that support bf16
do_eval=False,
# evaluation_strategy="no",
gradient_accumulation_steps=1,
gradient_checkpointing=True,
gradient_checkpointing_kwargs={"use_reentrant": False},
learning_rate=1.0e-04,
log_level="info",
logging_steps=5,
logging_strategy="steps",
lr_scheduler_type="cosine",
max_steps=-1,
num_train_epochs=5,
output_dir=output_dir,
overwrite_output_dir=True,
per_device_eval_batch_size=1,
per_device_train_batch_size=8,
save_strategy="no",
save_total_limit=None,
seed=42,
)
# based on config
peft_config = LoraConfig(
r=16,
lora_alpha=16,
lora_dropout=0.1,
bias="none",
task_type="CAUSAL_LM",
target_modules=["q_proj", "k_proj", "v_proj", "o_proj"],
)
trainer = SFTTrainer(
model=model_id,
model_init_kwargs=model_kwargs,
args=training_args,
train_dataset=train_dataset,
eval_dataset=train_dataset,
dataset_text_field="text",
tokenizer=tokenizer,
packing = True,
peft_config=peft_config,
max_seq_length=tokenizer.model_max_length,
)
train_result = trainer.train()
trainer.save_model(output_dir)
Now that we’ve completed the fine-tuning process, let’s now utilize the model for inference and obtain the performance metrics:
def parse_entities_from_trained_model(content):
"""
Extracts a list of entities from the output of a trained model.
Args:
- content (str): The raw string output from a trained model.
Returns:
- list of str: A list of entities extracted from the model's output.
"""
return content.split("The entities are:")[-1].split(",")
mistral_few_shot_answers = []
for item in tqdm(test_set_subsample):
few_shot_prompt_messages = build_entity_prompt(item)
# input_ids = tokenizer.apply_chat_template(few_shot_prompt_messages, tokenize=True, return_tensors = "pt").cuda()
prompt = tokenizer.apply_chat_template(few_shot_prompt_messages, tokenize=False)
tensors = tokenizer(prompt, return_tensors="pt")
input_ids = tensors.input_ids.cuda()
attention_mask = tensors.attention_mask.cuda()
outputs = model.generate(input_ids = input_ids, attention_mask = attention_mask, max_new_tokens=200, do_sample=False)
# https://github.com/huggingface/transformers/issues/17117#issuecomment-1124497554
gen_text = tokenizer.batch_decode(outputs.detach().cpu().numpy()[:, input_ids.shape[1]:], skip_special_tokens=True)[0]
mistral_few_shot_answers.append(parse_entities_from_trained_model(gen_text.strip()))
parsed_entities_few_shot = [[y["text"] for y in x] for x in mistral_few_shot_answers]
retrieved_answers = []
for item in tqdm(parsed_entities_few_shot):
answer_element = []
for entity in item:
retrieved_mesh_ids = entity_ranker.query(entity, top_n = 1)
answer_element.append({"identifier":retrieved_mesh_ids[0], "text":entity})
retrieved_answers.append(answer_element)
entity_scores = [calculate_entity_metrics(gt["annotations"],pred) for gt, pred in zip(test_set_subsample, retrieved_answers)]
macro_precision_entity = sum([x[0] for x in entity_scores]) / len(entity_scores)
macro_recall_entity = sum([x[1] for x in entity_scores]) / len(entity_scores)
macro_f1_entity = sum([x[2] for x in entity_scores]) / len(entity_scores)
mesh_scores = [calculate_mesh_metrics(gt["annotations"],pred) for gt, pred in zip(test_set_subsample, retrieved_answers)]
macro_precision_mesh = sum([x[0] for x in mesh_scores]) / len(mesh_scores)
macro_recall_mesh = sum([x[1] for x in mesh_scores]) / len(mesh_scores)
macro_f1_mesh = sum([x[2] for x in mesh_scores]) / len(mesh_scores)
This setup is exactly similar to the previous setup in that we continue to use the LLM as an entity extractor, and an external retriever for linking each entity to the MeSH ID. Fine-tuning the model leads to significant improvements across entity extraction and linking.
Compared to zero-shot entity extraction, fine-tuning improves all metrics by a factor of upto or more than 20%. Similarly, entity linking is also improved by a factor of around 12–14% across all metrics compared to the previous setting. These are not surprising takeaways though, as a task-specific model is expected to perform much better than the zero-shot performance. Still it’s nice to quantify these improvements concretely!
Benchmarking Scispacy
How does this implementation compare with an existing tool that can perform entity linking? Scispacy is a common work-horse for biomedical and clinical text processing, and provides features for entity extraction and entity linking. Specifically, Scispacy also provides a functionality to link entities to the MeSH KB, which is the file we also use as the KB originally for our LLM experiments. Let’s benchmark the performance of SciSpacy on our test set for comparison with our LLM experiments.
We use the “en_ner_bc5cdr_md” [15] in SciSpacy as the entity extraction module, as this model has been specifically trained on the BioCreative V dataset. Let’s evaluate the performance:
from scispacy.linking import EntityLinker
import spacy, scispacy
import pandas as pd
from helpers import *
from tqdm import tqdm
#code for setting up MeSH linker referred from https://github.com/allenai/scispacy/issues/355
config = {
"resolve_abbreviations": True,
"linker_name": "mesh",
"max_entities_per_mention":1
}
nlp = spacy.load("en_ner_bc5cdr_md")
nlp.add_pipe("scispacy_linker", config=config)
linker = nlp.get_pipe("scispacy_linker")
def extract_mesh_ids(text):
mesh_entity_pairs = []
doc = nlp(text)
for e in doc.ents:
if e._.kb_ents:
cui = e._.kb_ents[0][0]
mesh_entity_pairs.append({"text": e.text, "identifier": cui})
else:
mesh_entity_pairs.append({"text": e.text, "identifier": "None"})
return mesh_entity_pairs
all_mesh_ids = []
for item in tqdm(test_set_subsample):
text = item["title"] + " " + item["abstract"]
mesh_ids = extract_mesh_ids(text)
all_mesh_ids.append(mesh_ids)
entity_scores = [calculate_entity_metrics(gt["annotations"],pred) for gt, pred in zip(test_set_subsample, all_mesh_ids)]
macro_precision_entity = sum([x[0] for x in entity_scores]) / len(entity_scores)
macro_recall_entity = sum([x[1] for x in entity_scores]) / len(entity_scores)
macro_f1_entity = sum([x[2] for x in entity_scores]) / len(entity_scores)
mesh_scores = [calculate_mesh_metrics(gt["annotations"],pred) for gt, pred in zip(test_set_subsample, all_mesh_ids)]
macro_precision_mesh = sum([x[0] for x in mesh_scores]) / len(entity_scores)
macro_recall_mesh = sum([x[1] for x in mesh_scores]) / len(entity_scores)
macro_f1_mesh = sum([x[2] for x in mesh_scores]) / len(entity_scores)
Scispacy outperforms the fine-tuned LLM on entity extraction by a factor of 10% across all metrics, and by a factor of 14–20% on entity linking! For the task of biomedical entity extraction and linking, Scispacy remains a robust tool.
Takeaways
Having come to the end of our experiments, what are the concrete takeaways from them?
- Strengths in Zero-Shot Entity Extraction: Mistral-Instruct is a decent zero-shot entity extractor for biomedical text. While its parametric knowledge is not sufficient for performing zero-shot MeSH entity linking, we leverage it as an entity extractor in conjunction with an external KB retriever in our experiments to get much better performance.
- RAG’s Improvement over Zero-Shot Prediction: The LLM in a RAG setup demonstrates an improvement over a purely zero-shot approach for entity linking. However, the retriever component within a RAG setup can be a significant bottleneck, as in our case, the BM-25 retriever only manages to retrieve around 12–17% of relevant IDs per data point. This suggests a need for more effective retrieval methods.
- Pipelined extraction provides the best performance: Given the capabilities of the LLM as an entity extractor, the best performance is achieved when leveraging these capabilities within a larger pipeline that includes an external retriever to link entities to the MeSH knowledge base (KB). This is identical to the traditional setting, where entity extraction and KB-linking are kept as separate modules.
- Benefits of Fine-Tuning: Fine-tuning the LLM using QLora for the entity extraction task leads to significant performance improvements on entity extraction and entity linking when used in tandem with an external retriever.
- Scispacy performs the best: Scispacy outperforms all LLM-based methods for entity linking tasks in our experiments. For biomedical text processing, Scispacy remains a robust tool. It also requires less computational power for running compared to an LLM, which needs a good GPU for fast inference. In contrast, Scispacy only requires a good CPU.
- Opportunities for Optimization: Our current implementations of LLM-based pipelines for entity linking are quite naive with substantial room for improvement. Some areas that could benefit from optimization include the choice of retrieval and the retrieval logic itself. Fine-tuning the LLM with more data could also further boost its entity extraction performance.
Limitations
There are some limitations to our experiments so far.
- Multiple MeSH IDs for an entity: In our dataset, a few entities in each document could be linked to multiple MeSH IDs. Out of a total of 968 entities across 100 documents in our test set, this occurs in 15 cases (1.54%). In the Scispacy evaluation, as well as in all LLM experiments where we used the external KB linker (BM-25 retriever) after entity extraction, we link only one MeSH concept per entity. Although Scispacy offers the possibility of linking more than one MeSH ID per entity, we opt not to use this feature to ensure a fair comparison with our LLM experiments. Extending the functionality to support linking to more than one concept would also be an interesting addition.
- MeSH IDs not in the Knowledge Base: In the test dataset, there are MeSH IDs for entities that are not included in KB. Specifically, 64 entities (6.6% of cases) possess a MeSH ID that is absent from our KB. This limitation lies on the retriever side and can be addressed by updating the KB.
- Entities lacking a MeSH ID: Similarly, another 1.65% of entities (16 out of 968) cannot be mapped to a MeSH ID. In all LLM experiments where we use the external KB linker after entity extraction, we currently lack the ability to determine whether an entity has no MeSH ID.
References
I’ve included all papers and resources referred in this article here. Please let me know if I missed out on anything, and I will add them!
[1] Bodenreider O. (2004). The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic acids research, 32(Database issue), D267–D270. https://doi.org/10.1093/nar/gkh061
[2] https://www.nlm.nih.gov/healthit/snomedct/index.html
[3] https://www.nlm.nih.gov/mesh/meshhome.html
[4] Wei CH, Peng Y, Leaman R, Davis AP, Mattingly CJ, Li J, Wiegers TC, Lu Z. Overview of the BioCreative V Chemical Disease Relation (CDR) Task, Proceedings of the Fifth BioCreative Challenge Evaluation Workshop, p154–166, 2015
[5] Li J, Sun Y, Johnson RJ, Sciaky D, Wei CH, Leaman R, Davis AP, Mattingly CJ, Wiegers TC, Lu Z. Anotating chemicals, diseases and their interactions in biomedical literature, Proceedings of the Fifth BioCreative Challenge Evaluation Workshop, p173–182, 2015
[6] Leaman R, Dogan RI, Lu Z. DNorm: disease name normalization with pairwise learning to rank, Bioinformatics 29(22):2909–17, 2013
[7] Leaman R, Wei CH, Lu Z. tmChem: a high performance approach for chemical named entity recognition and normalization. J Cheminform, 7:S3, 2015
[8] Li, J., Sun, Y., Johnson, R. J., Sciaky, D., Wei, C. H., Leaman, R., Davis, A. P., Mattingly, C. J., Wiegers, T. C., & Lu, Z. (2016). BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database : the journal of biological databases and curation, 2016, baw068. https://doi.org/10.1093/database/baw068
[9] Jiang, A. Q., Sablayrolles, A., Mensch, A., Bamford, C., Chaplot, D. S., Casas, D. D. L., … & Sayed, W. E. (2023). Mistral 7B. arXiv preprint arXiv:2310.06825.
[10] Neumann, M., King, D., Beltagy, I., & Ammar, W. (2019, August). ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing. In Proceedings of the 18th BioNLP Workshop and Shared Task (pp. 319–327).
[11] https://ai2-s2-scispacy.s3-us-west-2.amazonaws.com/data/kbs/2020-10-09/mesh_2020.jsonl
[12] Lewis, P., Perez, E., Piktus, A., Petroni, F., Karpukhin, V., Goyal, N., … & Kiela, D. (2020). Retrieval-augmented generation for knowledge-intensive nlp tasks. Advances in Neural Information Processing Systems, 33, 9459–9474.
[13] Liu, N. F., Lin, K., Hewitt, J., Paranjape, A., Bevilacqua, M., Petroni, F., & Liang, P. (2024). Lost in the Middle: How Language Models Use Long Contexts. Transactions of the Association for Computational Linguistics, 12.
[14] Dettmers, T., Pagnoni, A., Holtzman, A., & Zettlemoyer, L. (2024). Qlora: Efficient finetuning of quantized llms. Advances in Neural Information Processing Systems, 36.
[15] https://s3-us-west-2.amazonaws.com/ai2-s2-scispacy/releases/v0.4.0/en_ner_bc5cdr_md-0.4.0.tar.gz
Building a Biomedical Entity Linker with LLMs was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
Originally appeared here:
Building a Biomedical Entity Linker with LLMs
Go Here to Read this Fast! Building a Biomedical Entity Linker with LLMs